The package is an appropriate way to organize the work and share it with others. Typically, a package will include code (not only R code!), documentation for the package and the functions inside, some tests to check everything works as it should, and data sets.
Packages in R
Packages in R Programming language are a set of R functions, compiled code, and sample data. These are stored under a directory called “library” within the R environment. By default, R installs a group of packages during installation. Once we start the R console, only the default packages are available by default. Other packages that are already installed need to be loaded explicitly to be utilized by the R program that’s getting to use them.
What are Repositories?
A repository is a place where packages are located and stored so you can install R packages from it. Organizations and Developers have a local repository, typically they are online and accessible to everyone. Some of the most popular repositories for R packages are:
- CRAN: Comprehensive R Archive Network(CRAN) is the official repository, it is a network of FTP and web servers maintained by the R community around the world. The R community coordinates it, and for a package to be published in CRAN, the Package needs to pass several tests to ensure that the package is following CRAN policies.
- Bioconductor: Bioconductor is a topic-specific repository, intended for open source software for bioinformatics. Similar to CRAN, it has its own submission and review processes, and its community is very active having several conferences and meetings per year in order to maintain quality.
- Github: Github is the most popular repository for open-source projects. It’s popular as it comes from the unlimited space for open source, the integration with git, a version control software, and its ease to share and collaborate with others.
Get library locations containing R packages
Output:
[1] "C:/Users/GFG19565/AppData/Local/Programs/R/R-4.3.1/library"
This method .libpath() might be related to managing library paths. Library paths are directories where a program looks for external libraries or modules that it needs to import or link during execution.
Get the list of all the R packages installed
Output:
Packages in library ‘C:/Users/GFG19565/AppData/Local/Programs/R/R-4.3.1/library’:
abind Combine Multidimensional Arrays
ade4 Analysis of Ecological Data: Exploratory and Euclidean
Methods in Environmental Sciences
askpass Password Entry Utilities for R, Git, and SSH
backports Reimplementations of Functions Introduced Since R-3.0.0
base The R Base Package
base64enc Tools for base64 encoding
BBmisc Miscellaneous Helper Functions for B. Bischl
BH Boost C++ Header Files
BiocManager Access the Bioconductor Project Package Repository
bit Classes and Methods for Fast Memory-Efficient Boolean
Selections
bit64 A S3 Class for Vectors of 64bit Integers
bitops Bitwise Operations
blob A Simple S3 Class for Representing Vectors of Binary
Data ('BLOBS')
boot Bootstrap Functions (Originally by Angelo Canty for S)
brew Templating Framework for Report Generation
brio Basic R Input Output
broom Convert Statistical Objects into Tidy Tibbles
bslib Custom 'Bootstrap' 'Sass' Themes for 'shiny' and
'rmarkdown'
cachem Cache R Objects with Automatic Pruning
callr Call R from R
car Companion to Applied Regression
carData Companion to Applied Regression Data Sets
caret Classification and Regression Training
caTools Tools: Moving Window Statistics, GIF, Base64, ROC AUC,
etc
We load a package using library(), the functions and objects in that package become available in the global environment.
Install an R-Packages
There are multiple ways to install R Package, some of them are,
- Installing R Packages From CRAN: For installing R Package from CRAN we need the name of the package and use the following command:
install.packages("package name")
- Installing Package from CRAN is the most common and easiest way as we just have to use only one command. In order to install more than a package at a time, we just have to write them as a character vector in the first argument of the install.packages() function:
Example:
R
install.packages(c("vioplot", "MASS"))
|
- Installing BiocManager Packages: In Bioconductor,using R version 3.5 or greater, which is not compatible with the biocLite.R script for installing Bioconductor packages.
Instead, you should use the BiocManager package to install and manage Bioconductor packages. Here’s an example of how to install the BiocManager package and use it to install a Bioconductor package.
R
install.packages("BiocManager")
|
This will install some basic functions which are needed to install bioconductor packages, such as the biocLite() function. To install the core packages of Bioconductor just type it without further arguments:
BiocManager::install("edgeR")
- This will install the edgeR package and its dependencies from the Bioconductor repository.
Update, Remove and Check Installed Packages in R
To check what packages are installed on your computer, type this command:
installed.packages()
To update all the packages, type this command:
update.packages()
To update a specific package, type this command:
install.packages("PACKAGE NAME")
Installing Packages Using RStudio UI
In R Studio goto Tools -> Install Package, and there we will get a pop-up window to type the package you want to install:
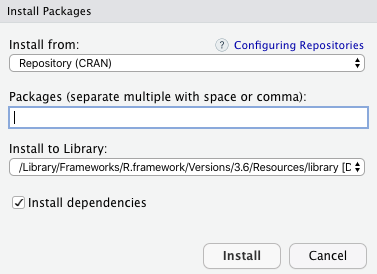
Packages in R Programming
Under Packages, type, and search Package which we want to install and then click on install button.
How to Load Packages in R Programming Language
When a R package is installed, we are ready to use its functionalities. If we just need a sporadic use of a few functions or data inside a package we can access them with the following notation.
# Load a package using the library function
library(dplyr)
# Load a package using the require function
require(dplyr)
Both functions attempt to load the specified package, but there is a subtle difference between the two: library() returns an error if the package is not found or cannot be loaded, whereas require() returns a warning and sets the value of the package variable to FALSE.
Difference Between a Package and a Library
There is always confusion between a package and a library, and we find people calling libraries as packages.
- library(): It is the command used to load a package, and it refers to the place where the package is contained, usually a folder on our computer.
- Package: It is a collection of functions bundled conveniently. The package is an appropriate way to organize our own work and share it with others.
Load More Than One Package at a Time
We can just input a vector of names to the install.packages() function to install an R package, in the case of the library() function, this is not possible. We can load a set of packages one at a time, or if you prefer, use one of the many workarounds developed by R users.
In R, you can load more than one package at a time using the library() function. Simply provide the names of the packages you want to load as a vector inside the library() function. Here’s an example:
# Load multiple packages at once
library(caret, dplyr, ggplot2)
This code loads the caret, dplyr, and ggplot2 packages at once.
Alternatively, you can use the require() function to load multiple packages, but this requires calling the function multiple times, once for each package:
# Load multiple packages using the require function
require(caret)
require(dplyr)
require(ggplot2)
Both methods accomplish the same thing, so it’s mostly a matter of personal preference. The library() function is more concise when loading multiple packages at once, while require() provides more control and can be used to load packages conditionally.
To install an R packages from GitHub from devtools package.
First, you need to install devtools by running the following code:
R
install.packages("devtools")
|
Output
.png)
Packages in R Programming
Once devtools is installed, you can use the install_github() function to install an R package from GitHub. The syntax is:
R
devtools::install_github("github_username/github_repo")
|
For example, to install the tidyverse package from GitHub, you would run:
R
devtools::install_github("tidyverse/tidyverse")
|
This will directly download and install the tidyverse package from GitHub.
It should be noted that certain packages may require the installation of extra dependencies before they can be utilised. In this instance, install_github() will attempt to install these dependencies as well. If you run into any problems during the installation, you may try manually installing any missing dependencies with the install.packages() function.
Choose the Right R Packages
The traditional way of discovering R packages is just by learning R, in many tutorials and courses the most popular packages are usually mentioned and used. The first alternative can be to browse categories of CRAN packages. CRAN is the official repository, also gives us the option to browse through packages.
Another alternative to finding packages can be R Documentation, a help documentation aggregator for R packages from CRAN, BioConductor, and GitHub, which offers you a search box ready for your requests directly on the main page.
Share your thoughts in the comments
Please Login to comment...