foreach parallel computing using external packages
Last Updated :
21 Aug, 2023
Parallel computing is a method of breaking down large computational tasks into smaller ones that can be executed simultaneously on multiple processors or cores, leading to faster results. The foreach loop is a popular construct in R programming, which allows users to iterate over a list or vector of elements. In parallel computing, foreach can be used to execute code in parallel across multiple processors or cores, leading to significant speedups in performance. In this article, we will discuss the concepts related to foreach parallel computing and the steps needed to use it, along with some good examples.
CONCEPTS:
The concept behind parallel computing is to break down a large computational task into smaller sub-tasks and execute them simultaneously across multiple processors or cores. In R programming, parallel computing can be achieved using the parallel package, which provides support for multiple types of parallelism, including fork-based, socket-based, and cluster-based parallelism.
The foreach loop is another popular construct in R programming that allows users to iterate over a list or vector of elements. The foreach package provides support for parallel computing using foreach loops, which allows users to execute code in parallel across multiple processors or cores.
Steps:
To use foreach parallel computing in R, the following steps are needed:
- Install the required packages: The foreach and doParallel packages are required to use foreach parallel computing in R. They can be installed using the following commands:
R
install.packages("foreach")
install.packages("doParallel")
|
- Load the packages: Once the packages are installed, they need to be loaded into the R environment using the following commands:
R
library(foreach)
library(doParallel)
|
- Create a cluster: Before executing code in parallel, a cluster needs to be created to specify the number of processors or cores to use. This can be done using the following command:
- Register the cluster: The cluster needs to be registered with the doParallel package using the following command:
- Execute code in parallel: Finally, the code can be executed in parallel using the foreach loop. The following code demonstrates how to calculate the sum of squares of a vector in parallel:
R
vec <- c(1:1000)
result <- foreach(i = 1:length(vec), .combine = "+") %dopar% {
vec[i]^2
}
|
In this example, the %dopar% operator is used to indicate that the code should be executed in parallel. The combine argument specifies the method to combine the results of each iteration, which in this case is the sum.
EXAMPLES:
The foreach package can be used in various scenarios where there is a need to iterate over a list or vector of elements and execute code in parallel. Some good examples include:
- Parallelizing a for loop : In R, the for loop is a popular construct used to iterate over a sequence of values. However, when dealing with large datasets, a for loop can take a significant amount of time to execute. By using the foreach package, we can parallelize the for loop and execute the iterations in parallel. The following code demonstrates how to do this:
R
library(foreach)
library(doParallel)
cl <- makeCluster(4)
registerDoParallel(cl)
vec <- 1:100
result <- foreach(i = 1:length(vec), .combine = c) %dopar% {
vec[i] * 2
}
print(result)
|
OUTPUT :

output
In this example, we create a cluster with 4 cores, register the cluster with the doParallel package, and then use foreach to parallelize the for loop. The code calculates the product of each value in the vector vec by 2, and stores the results in the variable result.
- Squaring numbers in parallel :-Suppose we have a vector of numbers and we want to square each number in parallel. Here’s how we can do it with foreach:
R
library(foreach)
library(doParallel)
cl <- makeCluster(4)
registerDoParallel(cl)
vec <- 1:10
result <- foreach(i = 1:length(vec), .combine = c) %dopar% {
vec[i]^2
}
stopCluster(cl)
print(result)
|
The 1:100 notation produces an integer vector from 1 to 100, which is a vector of values.The for loop that comes after it is parallelized using the foreach() method. The loop loops through the values in the vector, multiplying each value by two as it goes. The outcomes of the parallel calculations are combined into a single vector using the.combine = c argument.The foreach() loop uses the %dopar% operator to indicate that the loop should run concurrently on all available cores.The result variable holds the output of the concurrent computation.The computation’s findings are shown using the print() function.
OUTPUT :
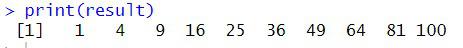
output
This code creates a cluster with 4 cores, defines a vector of numbers, and then parallelizes the for loop to square each number using the %dopar% operator. The .combine = c argument tells foreach to combine the results into a single vector. Finally, the code stops the cluster and prints the result.
- Finding the maximum of a list of matrices:
Suppose we have a list of matrices and we want to find the maximum value across all the matrices in parallel. Here’s how we can do it with foreach:
R
library(foreach)
library(doParallel)
cl <- makeCluster(4)
registerDoParallel(cl)
lst <- list(matrix(1:9, ncol = 3), matrix(10:18, ncol = 3), matrix(19:27, ncol = 3))
result <- foreach(mat = lst, .combine = max) %dopar% {
max(mat)
}
stopCluster(cl)
print(lst)
print(result)
|
OUTPUT :

output
This code creates a cluster with 4 cores, defines a list of matrices, and then parallelizes the for loop to find the maximum value across all the matrices using the %dopar% operator. The .combine = max argument tells foreach to combine the results using the max function. Finally, the code stops the cluster and prints the result.
- Parallel Matrix Multiplication:
This example demonstrates how to use foreach to parallelize matrix multiplication:
R
library(foreach)
library(doParallel)
cl <- makeCluster(4)
registerDoParallel(cl)
A <- matrix(rnorm(10000), 5, 5)
B <- matrix(rnorm(10000), 5, 5)
result <- foreach(i = 1:5, .combine = "cbind") %:%
foreach(j = 1:5, .combine = "c") %dopar% {
sum(A[i,] * B[,j])
}
stopCluster(cl)
print(A)
print(B)
print(result)
|
OUTPUT:
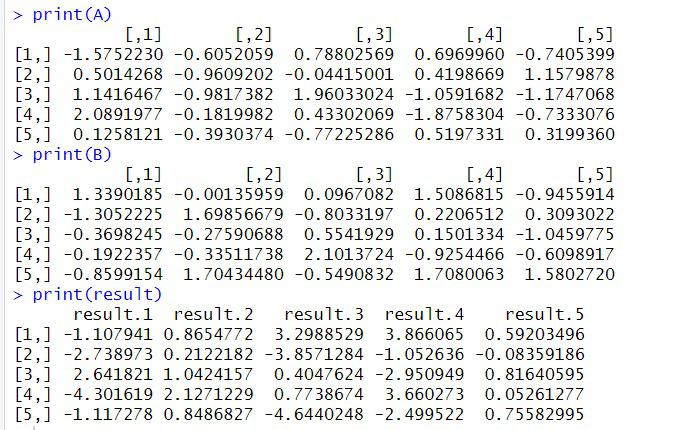
output
In this example, we create a cluster with 4 cores using makeCluster and register it for use with foreach. We then define two matrices A and B, and parallelize the matrix multiplication using foreach. The %:% operator indicates that the loops should be executed in parallel, and the .combine parameter specifies that the results should be combined using cbind and c to construct the resulting matrix. Finally, we stop the cluster and print the result
CONCLUSION:
In conclusion, foreach and doParallel are powerful R packages that enable users to parallelize their code and speed up computation on multi-core processors. By splitting a task into smaller chunks and distributing those chunks across multiple cores, users can dramatically reduce the time it takes to run computationally intensive code.
While parallel computing can be a powerful tool, it is important to keep in mind that it is not always the best solution for every problem. In some cases, parallelizing code can actually slow down computation due to overhead associated with distributing and combining results. Additionally, not all algorithms can be parallelized effectively, so it is important to carefully consider the nature of the problem and the structure of the code before attempting to parallelize.
Overall, foreach and doParallel are valuable tools to have in your R toolkit when working with computationally intensive code, and can help to significantly reduce the time it takes to perform complex simulations and data analysis.
Share your thoughts in the comments
Please Login to comment...