Local Tangent Space Alignment (LTSA) is a powerful dimensionality reduction technique used in machine learning and data analysis. In this article, we will provide a beginner-friendly introduction to the concept of LTSA, explain its primary terminologies, and guide you through the steps of implementing LTSA using Python’s scikit-learn library.
Local Tangent Space Alignment
A manifold learning technique called Local Tangent Space Alignment (LTSA) concentrates on maintaining local geometric correlations in high-dimensional data. The Low-dimensional Structural Analysis (LTSA) method creates a low-dimensional representation of the dataset by matching the tangent spaces of close data points. This method is useful in domains like computer vision and pattern recognition because it is very good at revealing hidden patterns in intricate datasets. When it comes to exposing the underlying geometry of data and offering insights into the fundamental structure of multidimensional information, LTSA is highly effective.
The goal of dimensionality reduction is to decrease the number of variables or features in a dataset while maintaining its organization and key information. Machine learning, data compression, noise reduction, and data visualization may all benefit from dimensionality reduction. Principal component analysis (PCA), linear discriminant analysis (LDA), and t-distributed stochastic neighbor embedding (t-SNE) are just a few of the several techniques available for reducing dimensionality. These techniques can only capture the linear correlations between the characteristics, though, since they are linear. These techniques might not be able to maintain the intrinsic geometry of the data if it contains nonlinear structures like curves, loops, or manifolds.
Manifold learning techniques, which are based on the notion that the high-dimensional data sits on or close to a low-dimensional manifold—a smooth surface that may be bent or twisted in the high-dimensional space—have been created to get around this restriction. Manifold learning techniques look for a low-dimensional data representation that maintains the angles or distances on the manifold.
In this article, we will explain the concept of LTSA, and show how to use the Sklearn library to implement it in Python. We will also demonstrate the results of LTSA on some synthetic and real-world datasets.
Key Terminologies of Local Tangent Space Alignment
Before we dive into the implementation of LTSA, let’s define some primary terminologies:
- Local Tangent Space: This is a local neighborhood around a data point where we compute the tangent space. It captures the local structure of the data.
- Manifold: A manifold is a smooth, linked, lower-dimensional subset of a high-dimensional space that contains data points. The LTSA is looking for this manifold.
- Local Linearity: LTSA assumes that data points on the manifold are locally linear and focuses on preserving these local linear relationships.
- Embedding: The lower-dimensional representation of data after applying LTSA.
Now, let’s proceed to implement LTSA using scikit-learn.
How LTSA works?
The primary goal of LTSA is to use a linear subspace at each location, known as the tangent space, to approximate the local geometry of the manifold. The plane that fits the neighboring points on the manifold the best is called the tangent space. Next, to create a global coordinate system that maintains the local angles and distances, LTSA attempts to align these tangent spaces.
The following are the LTSA steps:
- Determine the k closest neighbors of each point x in the dataset, then create a neighborhood matrix X for each point. The initial dimension of the data was d, and the matrix X had k rows and d columns.
- Take the mean of each column and remove it to center the neighborhood matrix X. Next, take the centered matrix X and apply Singular Value Decomposition (SVD) to get the singular values S, the right singular vectors V, and the left singular vectors U. There are three types of matrices: d rows and k columns in matrix V, k rows and k columns in matrix S, and a diagonal matrix with k elements in matrix U.
- Choose the first n columns of U, where n is the required reduced data dimension. The neighborhood points’ local coordinates in the tangent space are formed by these columns. There are k rows and n columns in the matrix U.
- To obtain a collection of local coordinate matrices U, repeat steps 1 through 3 for each point in the dataset. Next, create a sparse matrix W of the same size as the data matrix X, where the weight of each edge from point
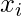 to point
to point  is represented by the element
is represented by the element  . In the event where
. In the event where  is not a neighbor of
is not a neighbor of 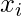 , the weight
, the weight  is equal to the corresponding element in the local coordinate matrix U; otherwise, it is equal to zero.
is equal to the corresponding element in the local coordinate matrix U; otherwise, it is equal to zero. - For the matrix
 where I is the identity matrix, find the eigenvalues and eigenvectors. The matrix M is positive semi-definite, symmetric, and contains d rows and d columns. The eigenvalues determine the order in which the eigenvectors and eigenvalues are arranged.
where I is the identity matrix, find the eigenvalues and eigenvectors. The matrix M is positive semi-definite, symmetric, and contains d rows and d columns. The eigenvalues determine the order in which the eigenvectors and eigenvalues are arranged. - After removing the zero eigenvalues, choose the n eigenvectors that match the fewest n eigenvalues. The global coordinates of the data points in the reduced space are formed by these eigenvectors. There are d rows and n columns in the matrix Y.
Implementation of LTSA in Sklearn
Sklearn is a popular Python library for machine learning and data analysis. It provides various tools and algorithms for data preprocessing, feature extraction, dimensionality reduction, clustering, classification, regression, and model evaluation.
Sklearn also implements several manifold learning methods, including LTSA. To use LTSA in Sklearn, we need to import the LocallyLinearEmbedding class from the sklearn. manifold module, and specify the method parameter as ‘ltsa’. The other parameters are:
- n_neighbors: the number of neighbors to consider for each point. The default value is 5.
- n_components: the number of dimensions for the reduced data. The default value is 2.
- reg: the regularization constant, which multiplies the trace of the local covariance matrix of the distances. The default value is 0.001.
- eigen_solver: the solver used to compute the eigenvectors. The available options are ‘auto’, ‘arpack’, and ‘dense’. The default value is ‘auto’.
- tol: the tolerance for the ‘arpack’ solver. The default value is 1e-6.
- max_iter: the maximum number of iterations for the ‘arpack’ solver. The default value is 100.
- random_state: the seed for the random number generator. The default value is None.
The LocallyLinearEmbedding class has a fit_transform method, which takes the data matrix X as input, and returns the reduced matrix Y as output. The class also has an attribute reconstruction_error_, which stores the reconstruction error associated with the embedding.
The following code shows how to use LTSA in Sklearn on a synthetic dataset:
Implementation of Local Tangent Space Alignment using Synthetic dataset
We will use a synthetic dataset of 1000 points sampled from a Swiss roll surface. This dataset has three dimensions, but only two are informative. We will use LTSA to reduce the dimensionality to two and visualize the result.
Import the necessary libraries
Python
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import make_swiss_roll
from sklearn.manifold import LocallyLinearEmbedding
np.random.seed(42)
|
This code sample uses scikit-learn’s make_swiss_roll to create a synthetic 3D dataset that resembles a swiss roll. Next, in order to decrease the dimensionality of the data while maintaining its local geometric structure, it employs locally linear embedding, or LLE. Lastly, a 2D visualization of the original and embedded data is produced using matplotlib. To guarantee repeatability, the random seed is set.
Generate the synthetic dataset using the make_swiss_roll
Python
X, color = make_swiss_roll(n_samples=1000, noise=0.1)
|
This function returns two arrays: the first one contains the coordinates of the points, and the second one contains the univariate position of the points on the manifold.
Plot the original dataset in 3D
We will use the color array to indicate the position of the points on the manifold.
Python
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(X[:, 0], X[:, 1], X[:, 2], c=color, cmap=plt.cm.Spectral)
ax.set_title('Original dataset')
plt.show()
|
Output:
.jpg)
This code takes a 3D dataset called X and uses matplotlib to build a 3D scatter plot. For the X, Y, and Z axes, respectively, each point’s coordinates from the first, second, and third dimensions are utilized. The color array dictates the color of the points, and the plot is shown with the title “Original dataset.”
Apply LTSA to the dataset using the LocallyLinearEmbedding
Python
ltsa = LocallyLinearEmbedding(n_components=2, method='ltsa', n_neighbors=10)
X_ltsa = ltsa.fit_transform(X)
|
We will use the n_components parameter to specify the number of dimensions we want to reduce to, and the method parameter to specify the LTSA algorithm. We will also use the n_neighbors parameter to specify the number of nearest neighbors to use for each point. This parameter affects the local structure of the manifold. We will use 10 neighbors as a reasonable choice.
Plot the transformed dataset
We will use the same color array as before to indicate the position of the points on the manifold.
Python
plt.figure(figsize=(8, 6))
plt.scatter(X_ltsa[:, 0], X_ltsa[:, 1], c=color, cmap=plt.cm.Spectral)
plt.title('LTSA result')
plt.show()
|
Output:
-(1).jpg)
This code uses matplotlib to create a 2D scatter plot for the data that has been locally linearly embedded (LTSA) processed. The X_ltsa file stores the reduced-dimensional data, and the Spectral colormap is used by the color array to determine each point’s color. The title of the plot is “LTSA result.”
We can see that LTSA has successfully unfolded the Swiss roll and preserved the local structure of the data. The points that are close on the manifold are also close in the lower-dimensional space.
Conclusion
In this article, we have explained the concept of Local Tangent Space Alignment (LTSA), which is a manifold learning method for dimensionality reduction. We have shown how to use the Sklearn library to implement LTSA in Python, and demonstrated the results of LTSA on some synthetic and real-world datasets. We have also discussed some of the advantages and limitations of LTSA, and compared it with other manifold learning methods. We hope that this article has given you a better understanding of LTSA and its applications in data analysis and visualization.
Share your thoughts in the comments
Please Login to comment...