Simple 1D Kernel Density Estimation in Scikit Learn
Last Updated :
08 Jun, 2023
In this article, we will learn how to use Scikit learn for generating simple 1D kernel density estimation. We will first understand what is kernel density estimation and then we will look into its implementation in Python using KernelDensity class of sklearn.neighbors in scikit learn library.
Kernel density estimation
A non-parametric method for estimating the probability density function of a continuous random variable using kernels as weights is known as kernel density estimation (or KDE). It is used in a variety of tasks, like data visualization, data analysis, and machine learning. The idea behind KDE is to treat each observation point as a small probability distribution, and the density estimate is obtained by aggregating these distributions.
A kernel function is employed in KDE to estimate the density at each data point, and the separate kernel estimates are then added together to provide the final density estimate. The predicted density’s shape is determined by the kernel function, a symmetric, smooth function that is typically bell-shaped.
The formula for KDE can be expressed as follows:
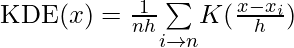
Where:
- KDE(x) is the estimated density at point x.
- n is the number of data points.
- h is the bandwidth, which controls the smoothness of the estimated density.
- xi represents each data point in the dataset.
- K(x;h) is the kernel function, which determines the contribution of each data point to the estimated density.
Some of the Commonly used kernel functions include:
- Gaussian (or Normal) Kernel: K(x,h) = \frac{1}{\sqrt{2\pi}}\exp\left[\frac{(x-x_i)^2}{2h^2}\right]
- Tophat Kernel: K(x;h) = \frac{1}{\sqrt{2\pi}}\exp\left(\frac{1}{2}\left|\frac{x-x_i}{h}\right|\right)
- Epanechnikov Kernel: K(x;h) = \left[\frac{3}{4}\left(1 – (\frac{x – x_i}{h})^2 (|\frac{x – x_i}{h}| \leq 1\right)\right]
- Exponential Kernel: K(x;h) = 0.5 \exp(-|\frac{x-x_i}{h}|)
- Linear Kernel:
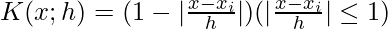
- Cosine Kernel: K(x;h) = \frac{\pi}{4}\cos( \frac{\pi}{2}\left(\frac{x-x_i}{h}\right)) (|\frac{x-x_i}{h}| \leq 1)
The kernels used in KDE are smooth, symmetric, and usually, bell-shaped functions, and the type of kernel and width of the kernel (bandwidth) determines the smoothness and accuracy of the estimated density. The estimated density at a particular point is obtained by calculating the weighted average of the kernel functions centered at each data point, with the weights determined by the distance between the point of interest and the data points.
Code Implementation
To implement this we will first import the required libraries.
Python3
import numpy as np
import matplotlib.pyplot as plt
from sklearn.neighbors import KernelDensity
|
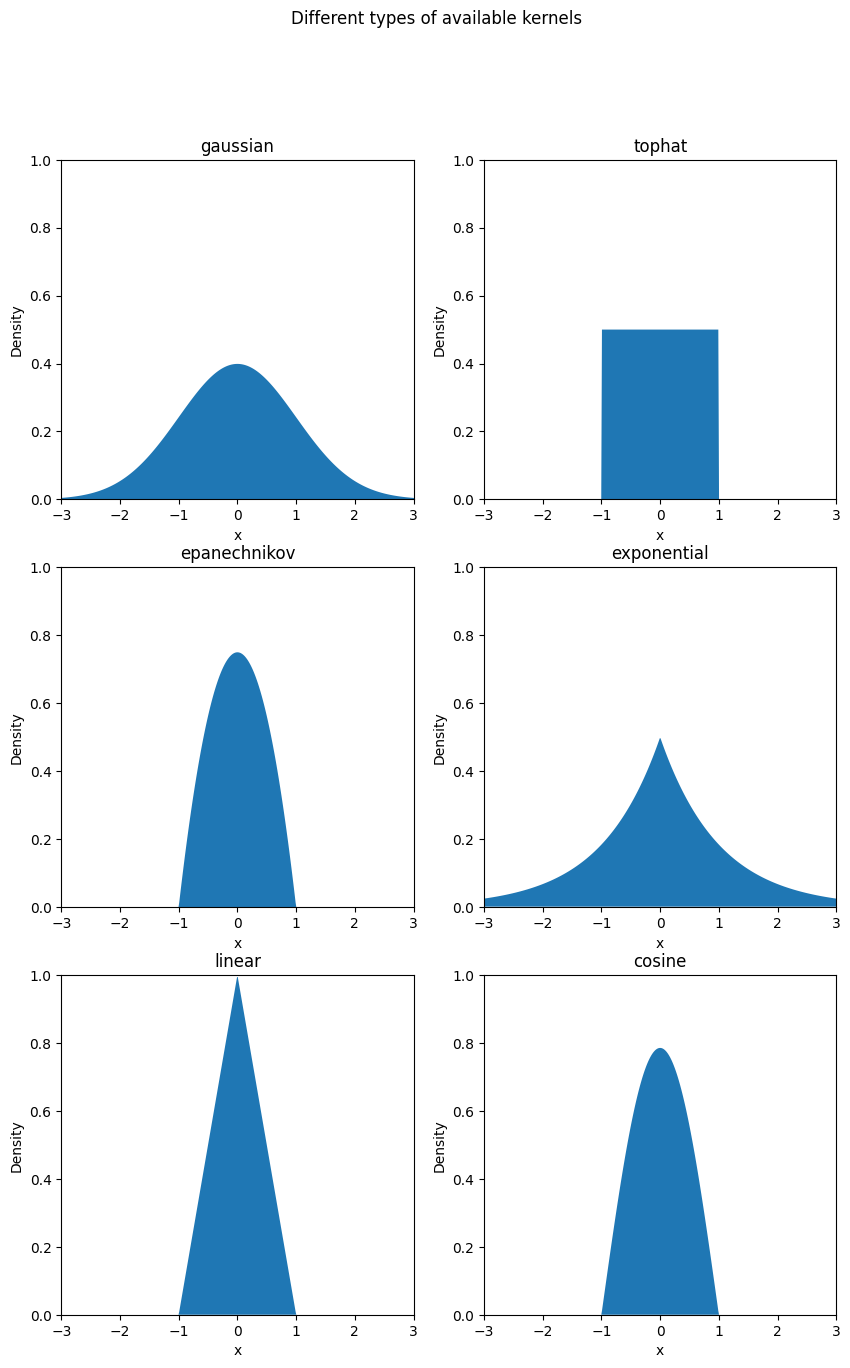
Now, we will look into different kernels available to use in this library.
Python3
kernels = ["gaussian", "tophat", "epanechnikov", "exponential", "linear", "cosine"]
fig, ax = plt.subplots(3, 2)
fig.set_figheight(15)
fig.set_figwidth(10)
fig.suptitle("Different types of available kernels")
x_plot = np.linspace(-6, 6, 1000)[:, np.newaxis]
x_src = np.zeros((1, 1))
for i, kernel in enumerate(kernels):
log_dens = KernelDensity(kernel=kernel).fit(x_src).score_samples(x_plot)
ax[i // 2, i % 2].fill(x_plot[:, 0], np.exp(log_dens))
ax[i // 2, i % 2].set_title(kernel)
ax[i // 2, i % 2].set_xlim(-3, 3)
ax[i // 2, i % 2].set_ylim(0, 1)
ax[i // 2, i % 2].set_ylabel("Density")
ax[i // 2, i % 2].set_xlabel("x")
plt.show()
|
Output:
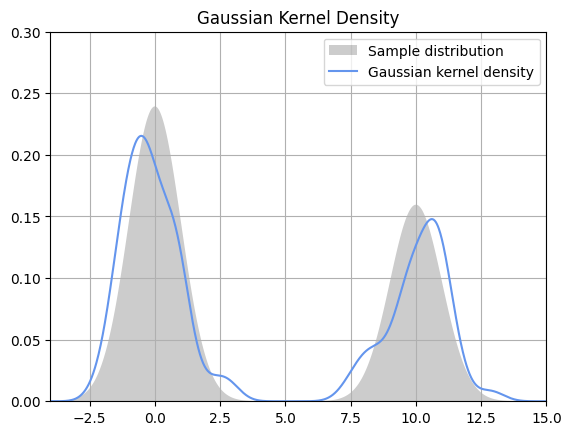
Now, let us implement kernel density estimation on random data using a Gaussian kernel with a bandwidth of 0.5.
Python3
N = 100
X = np.concatenate((np.random.normal(0, 1, int(0.6 * N)),
np.random.normal(10, 1, int(0.4 * N)))
)[:, np.newaxis]
X_plot = np.linspace(-5, 15, 1000)[:, np.newaxis]
true_density = 0.6 * norm(0, 1).pdf(X_plot[:, 0]) + \
0.4 * norm(10, 1).pdf(X_plot[:, 0])
fig, ax = plt.subplots()
ax.fill(
X_plot[:, 0], true_density,
fc='black', alpha=0.2,
label='Sample distribution'
)
kde = KernelDensity(kernel='gaussian', bandwidth=0.5).fit(X)
log_dens = kde.score_samples(X_plot)
ax.plot(
X_plot[:, 0],
np.exp(log_dens),
color="cornflowerblue",
linestyle="-",
label="Gaussian kernel density"
)
ax.set_title("Gaussian Kernel Density")
ax.set_xlim(-4, 15)
ax.set_ylim(0, 0.3)
ax.grid(True)
ax.legend(loc='upper right')
plt.show()
|
Output:
Share your thoughts in the comments
Please Login to comment...