Permutation Hypothesis Test in R Programming
Last Updated :
24 Nov, 2020
In simple words, the permutation hypothesis test in R is a way of comparing a numerical value of 2 groups. The permutation Hypothesis test is an alternative to:
Let’s implement this test in R programming.
Why use the Permutation Hypothesis Test?
- Small Sample Size.
- Assumptions(for parametric approach) not met.
- Test something other than classic approaches comparing Means and Medians.
- Difficult to estimate the SE for test-statistic.
Permutation Hypothesis Test Steps
- Specify a hypothesis
- Choose test-stat(Eg: Mean, Median, etc. )
- Determine Distribution of test-stat
- Convert test-stat to P-value
Note:
P-value = No. of permutations having a test-stat value greater than observed test-stat value/ No. of permutations.
Implementation in R
- Dataset: Chicken Diet Data. This dataset is a subset of the “chickwts” data in the “R dataset package”. Download the data set here.
- Hypothesis: The weight of the chicken is independent of the type of diet.
Test-Statistics
- Test-Statistics #1: The absolute value of the difference in mean weights for the two diets | Y1 – Y2 |. This is the same test statistics as the independent two-sided two-sample t-test.
- Test-Statistics #2: The absolute value of the difference in median weights for the two diets | Median1 – Median2 |
R
d <- read.table(file = "ChickData.csv",
header = T, sep = ",")
print(d)
names(d)
levels(d$feed)
table(d$feed)
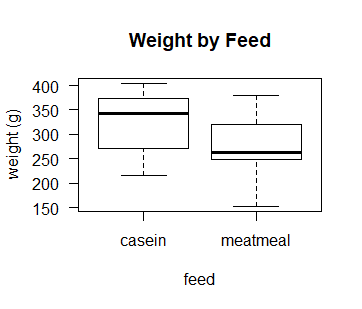
boxplot(d$weight~d$feed, las = 1,
ylab = "weight (g)",
xlab = "feed",
main = "Weight by Feed")
mean(d$weight[d$feed == "casein"])
mean(d$weight[d$feed == "meatmeal"])
test.stat1 <- abs(mean(d$weight[d$feed == "casein"]) -
mean(d$weight[d$feed == "meatmeal"]))
test.stat1
median(d$weight[d$feed == "casein"])
median(d$weight[d$feed == "meatmeal"])
test.stat2 <- abs(median(d$weight[d$feed == "casein"]) -
median(d$weight[d$feed == "meatmeal"]))
test.stat2
set.seed(1979)
n <- length(d$feed)
P <- 100000
variable <- d$weight
PermSamples <- matrix(0, nrow = n, ncol = P)
for(i in 1:P)
{
PermSamples[, i] <- sample(variable,
size = n,
replace = FALSE)
}
PermSamples[, 1:5]
Perm.test.stat1 <- Perm.test.stat2 <- rep(0, P)
for (i in 1:P)
{
Perm.test.stat1[i] <- abs(mean(PermSamples[d$feed == "casein",i]) -
mean(PermSamples[d$feed == "meatmeal",i]))
Perm.test.stat2[i] <- abs(median(PermSamples[d$feed == "casein",i]) -
median(PermSamples[d$feed == "meatmeal",i]))
}
test.stat1; test.stat2
round(Perm.test.stat1[1:15], 1)
round(Perm.test.stat2[1:15], 1)
(Perm.test.stat1 >= test.stat1)[1:15]
mean((Perm.test.stat1 >= test.stat1)[1:15])
mean(Perm.test.stat1 >= test.stat1)
mean(Perm.test.stat2 >= test.stat2)
|
Output:
> print(d)
weight feed
1 325 meatmeal
2 257 meatmeal
3 303 meatmeal
4 315 meatmeal
5 380 meatmeal
6 153 meatmeal
7 263 meatmeal
8 242 meatmeal
9 206 meatmeal
10 344 meatmeal
11 258 meatmeal
12 368 casein
13 390 casein
14 379 casein
15 260 casein
16 404 casein
17 318 casein
18 352 casein
19 359 casein
20 216 casein
21 222 casein
22 283 casein
23 332 casein
> names(d)
[1] "weight" "feed"
> levels(d$feed)
[1] "casein" "meatmeal"
> table(d$feed)
casein meatmeal
12 11
> mean(d$weight[d$feed == "casein"]) # mean for casein
[1] 323.5833
> mean(d$weight[d$feed == "meatmeal"]) # mean for meatmeal
[1] 276.9091
> test.stat1
[1] 46.67424
> median(d$weight[d$feed == "casein"]) # median for casein
[1] 342
> median(d$weight[d$feed == "meatmeal"]) # median for meatmeal
[1] 263
> test.stat2
[1] 79
> PermSamples[, 1:5]
[,1] [,2] [,3] [,4] [,5]
[1,] 379 283 380 352 206
[2,] 380 303 258 260 380
[3,] 257 206 379 380 153
[4,] 283 242 222 404 359
[5,] 222 260 325 258 258
[6,] 315 352 153 379 263
[7,] 352 263 263 325 325
[8,] 153 325 315 359 216
[9,] 368 379 344 242 260
[10,] 344 258 368 368 257
[11,] 359 257 206 257 315
[12,] 206 153 404 222 303
[13,] 404 344 303 390 390
[14,] 325 318 318 303 352
[15,] 242 404 332 263 404
[16,] 390 380 257 206 379
[17,] 260 332 216 315 318
[18,] 303 359 352 344 368
[19,] 263 222 242 283 222
[20,] 332 368 260 332 344
[21,] 318 315 283 318 283
[22,] 216 390 390 153 332
[23,] 258 216 359 216 242
> test.stat1; test.stat2
[1] 46.67424
[1] 79
> round(Perm.test.stat1[1:15], 1)
[1] 17.1 32.4 17.6 47.1 56.1 28.9 31.0 40.8 6.8 13.8 9.1 46.5 28.9 50.9 32.7
> round(Perm.test.stat2[1:15], 1)
[1] 61.0 75.0 4.5 59.0 78.0 17.0 62.0 38.5 4.5 16.0 23.0 60.5 63.5 75.0 37.0
> (Perm.test.stat1 >= test.stat1)[1:15]
[1] FALSE FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE
> mean((Perm.test.stat1 >= test.stat1)[1:15])
[1] 0.2
> mean(Perm.test.stat1 >= test.stat1)
[1] 0.09959
> mean(Perm.test.stat2 >= test.stat2)
[1] 0.05407
Share your thoughts in the comments
Please Login to comment...