While solving problems in real life it is very rare that we only come across binary classification problems because there are times when we have to classify within multiple categories for example dealing with the iris problem or the MNIST dataset is one of the common multiclass classification problems. As the researchers are developing the state of the art models they also keep this in mind to make the model work for multiclass classification problems.
Idea Behind using MultiClass Classification using LightGBM
As we know the lightGBM model is a light gradient-boosted machine that is used for the purpose of training on larger datasets in less time and getting decent results as well. As there are fewer models that support multiclass classification and the lightgbm model is one of them just we need to pass the right parameters to the model. We even do not have to convert it into the one hot encoded class it performs this on its own, which is an advantage here of using the LightGBM model for multi-class classification.
params = {
'objective': 'multiclass',
'num_class': 4,
'metric': 'multi_logloss',
'verbose': 0
}
For example in the above parameters:
- objective – We will set that to the multiclass which will tell teh model that we are trying to train a model on the multi-class classification dataset.
- metric – As the objective change we will have to change our metric as well we can use the multi_logloss, auc_mu or cross_entropy.
One more reason to use the LightGBM model is that it gives result comparative to the Neural Networks and as they are very efficient in tasks like this that is multi-class problems but implementing a NN for such a task can be tedious so, LightGBM comes handy and is easy to implement with some tweaks and twerks in the params of teh model.
Multiclass classification using LightGBM
In this article, we will learn about LightGBM model usage for the multiclass classification problem. This dataset has been used in this article to perform EDA on it and train the LightGBM model on this multiclass classification problem. But to use the LightGBM model we will first have to install the LightGBM model using the below command (in this article we are using version 3.3.5) :
!pip install lightgbm==3.3.5
Importing Libraries and Dataset
Python and its libraries are very helpful when given a large dataset which requires handling and processing.
- Pandas – This library is useful in loading the data and performing complex transformation.
- Numpy – Numpy arrays are useful when dealing with complex mathematical functions which require faster processing and performance
- Matplotlib/Seaborn – This library is useful in creating meaningful visualizations for better inference.
- Sklearn – This contains a vast library of useful functions which can be used for varied reasons like pre processing, predictions, etc.
Python3
import pandas as pd
import numpy as np
import seaborn as sb
import matplotlib.pyplot as plt
from sklearn.metrics import accuracy_score
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
import lightgbm as lgb
from sklearn.metrics import classification_report
import warnings
warnings.filterwarnings('ignore')
|
Now lets load the dataset into pandas dataframe.
Python3
df = pd.read_csv('bodyPerformance.csv')
df.head()
|
Output:
age gender height_cm weight_kg body fat_% diastolic systolic \
0 27.0 M 172.3 75.24 21.3 80.0 130.0
1 25.0 M 165.0 55.80 15.7 77.0 126.0
2 31.0 M 179.6 78.00 20.1 92.0 152.0
3 32.0 M 174.5 71.10 18.4 76.0 147.0
4 28.0 M 173.8 67.70 17.1 70.0 127.0
... ... ... ... ... ... ... ...
13388 25.0 M 172.1 71.80 16.2 74.0 141.0
13389 21.0 M 179.7 63.90 12.1 74.0 128.0
13390 39.0 M 177.2 80.50 20.1 78.0 132.0
13391 64.0 F 146.1 57.70 40.4 68.0 121.0
13392 34.0 M 164.0 66.10 19.5 82.0 150.0
gripForce sit and bend forward_cm sit-ups counts broad jump_cm class
0 54.9 18.4 60.0 217.0 C
1 36.4 16.3 53.0 229.0 A
2 44.8 12.0 49.0 181.0 C
3 41.4 15.2 53.0 219.0 B
4 43.5 27.1 45.0 217.0 B
... ... ... ... ... ...
13388 35.8 17.4 47.0 198.0 C
13389 33.0 1.1 48.0 167.0 D
13390 63.5 16.4 45.0 229.0 A
13391 19.3 9.2 0.0 75.0 D
13392 35.9 7.1 51.0 180.0 C
[13393 rows x 12 columns]
Output:
(13393, 12)
By using the df.info() function we can see the content of each columns and the data types present in it along with the number of null values present in each column.
Output:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 13393 entries, 0 to 13392
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 age 13393 non-null float64
1 gender 13393 non-null object
2 height_cm 13393 non-null float64
3 weight_kg 13393 non-null float64
4 body fat_% 13393 non-null float64
5 diastolic 13393 non-null float64
6 systolic 13393 non-null float64
7 gripForce 13393 non-null float64
8 sit and bend forward_cm 13393 non-null float64
9 sit-ups counts 13393 non-null float64
10 broad jump_cm 13393 non-null float64
11 class 13393 non-null object
dtypes: float64(10), object(2)
memory usage: 1.2+ MB
Descriptive statistical measures of the dataset help us better understand the data and it’s distribution over the plane. Let’s explore that to better understand it.
Output:
age height_cm weight_kg body fat_% diastolic \
count 13393.000000 13393.000000 13393.000000 13393.000000 13393.000000
mean 36.775106 168.559807 67.447316 23.240165 78.796842
std 13.625639 8.426583 11.949666 7.256844 10.742033
min 21.000000 125.000000 26.300000 3.000000 0.000000
25% 25.000000 162.400000 58.200000 18.000000 71.000000
50% 32.000000 169.200000 67.400000 22.800000 79.000000
75% 48.000000 174.800000 75.300000 28.000000 86.000000
max 64.000000 193.800000 138.100000 78.400000 156.200000
systolic gripForce sit and bend forward_cm sit-ups counts \
count 13393.000000 13393.000000 13393.000000 13393.000000
mean 130.234817 36.963877 15.209268 39.771224
std 14.713954 10.624864 8.456677 14.276698
min 0.000000 0.000000 -25.000000 0.000000
25% 120.000000 27.500000 10.900000 30.000000
50% 130.000000 37.900000 16.200000 41.000000
75% 141.000000 45.200000 20.700000 50.000000
max 201.000000 70.500000 213.000000 80.000000
broad jump_cm
count 13393.000000
mean 190.129627
std 39.868000
min 0.000000
25% 162.000000
50% 193.000000
75% 221.000000
max 303.000000
Exploratory Data Analysis
Exploratory Data analysis (EDA), as the name suggests, is used to “explore” about the data. This means that in EDA, we are given with the task to learn about what story/pattern the dataset exhibits. This can be sued to get trends out of the dataset, find hidden patterns, check our hypothesis and much more, both in visual and statistical manner.
Python3
plt.subplot(1, 2, 1)
temp = df['gender'].value_counts()
plt.pie(temp.values, labels=temp.index.values,
autopct='%1.1f%%')
plt.title("Distribution of Gender")
plt.subplot(1, 2, 2)
temp = df['class'].value_counts()
plt.pie(temp.values, labels=temp.index.values,
autopct='%1.1f%%')
plt.title("Class Distribution")
plt.show()
|
Output:
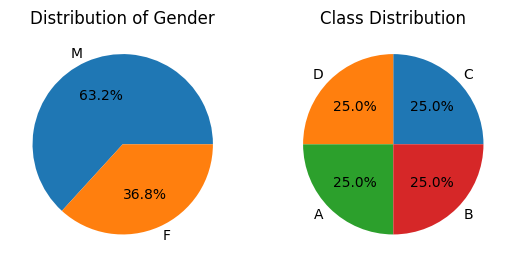
In the dataset we can observe that the most of the columns are the numerical ones. And to explore the continuous data density plot is the most preferred one. But to create density plot iteratively we will have to first segregate all the numerical columns.
Python3
num_cols = list()
for col in df.columns:
if df[col].dtype == 'object':
continue
num_cols.append(col)
num_cols
|
Output:
['age',
'height_cm',
'weight_kg',
'body fat_%',
'diastolic',
'systolic',
'gripForce',
'sit and bend forward_cm',
'sit-ups counts',
'broad jump_cm']
Now let’s create the density plot for all these numerical columns that we have segregated above.
Python3
plt.subplots(figsize=(8, 7))
for i, col in enumerate(num_cols):
plt.subplot(4, 3, i+1)
sb.distplot(df[col])
plt.tight_layout()
plt.show()
|
Output:
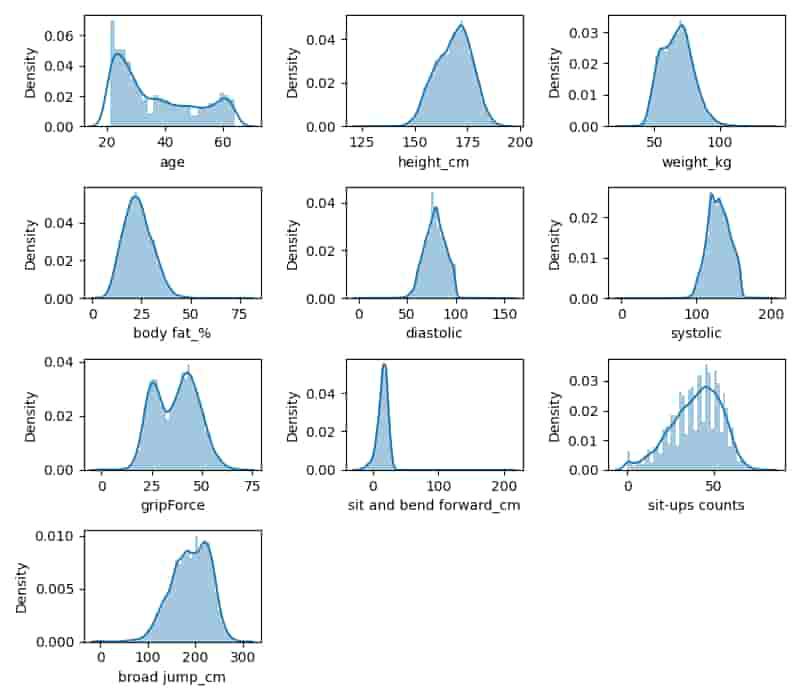
Distplot for various features
From the density plot above we can observe that the most of the numerical features are left skewed. And some of the features are normally distributed as well like BMI, Glucose, BloodPressure.
Python3
df['gender'] = df['gender'].map({'M': 0, 'F': 1})
df['class'] = df['class'].map({'A': 0, 'B': 1,
'C': 2, 'D': 3})
df.head()
|
Output:
age gender height_cm weight_kg body fat_% diastolic systolic \
0 27.0 0 172.3 75.24 21.3 80.0 130.0
1 25.0 0 165.0 55.80 15.7 77.0 126.0
2 31.0 0 179.6 78.00 20.1 92.0 152.0
3 32.0 0 174.5 71.10 18.4 76.0 147.0
4 28.0 0 173.8 67.70 17.1 70.0 127.0
... ... ... ... ... ... ... ...
13388 25.0 0 172.1 71.80 16.2 74.0 141.0
13389 21.0 0 179.7 63.90 12.1 74.0 128.0
13390 39.0 0 177.2 80.50 20.1 78.0 132.0
13391 64.0 1 146.1 57.70 40.4 68.0 121.0
13392 34.0 0 164.0 66.10 19.5 82.0 150.0
gripForce sit and bend forward_cm sit-ups counts broad jump_cm \
0 54.9 18.4 60.0 217.0
1 36.4 16.3 53.0 229.0
2 44.8 12.0 49.0 181.0
3 41.4 15.2 53.0 219.0
4 43.5 27.1 45.0 217.0
... ... ... ... ...
13388 35.8 17.4 47.0 198.0
13389 33.0 1.1 48.0 167.0
13390 63.5 16.4 45.0 229.0
13391 19.3 9.2 0.0 75.0
13392 35.9 7.1 51.0 180.0
class
0 2
1 0
2 2
3 1
4 1
... ...
13388 2
13389 3
13390 0
13391 3
13392 2
[13393 rows x 12 columns]
Finding Correlation between the features
Now we will find the correlation between the features in the dataset. It is to check that
Python3
sb.heatmap(df.corr(), annot=True,
cbar=True, cmap='RdBu', fmt='.1f')
plt.savefig('heatmap.png')
plt.show()
|
Output:
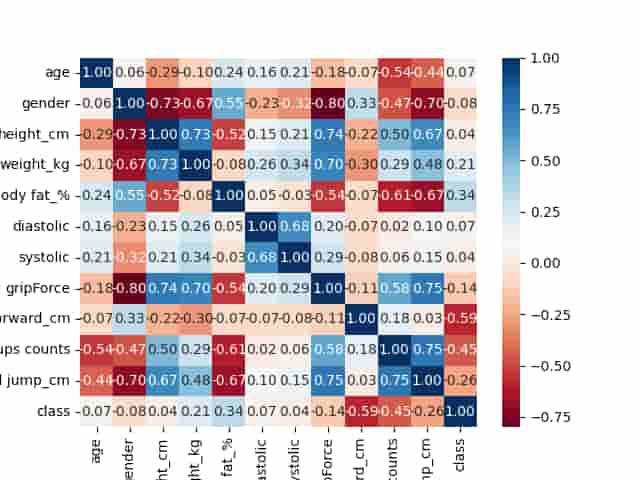
As there are no highly correlated feature (correlation greater than 0.8) in the dataset that can lead to data leakage or unnecessary increased complexity of the model, we can now move forward to data preprocessing and model development.
Model Development and Data Pre-processing
The first and the foremost step is that of splitting the data into training and the validation data. So, that we can monitor the training process of the model on some unforeseen data.
Python3
features = df.drop('class', axis=1)
target = df['class']
X_train, X_val,\
Y_train, Y_val = train_test_split(features, target,
random_state=2023,
test_size=0.20)
X_train.shape, X_val.shape
|
Output:
((10714, 11), (2679, 11))
Feature scaling proves to be very helpful in the fast and the stable training of the model.
Python3
scaler = StandardScaler()
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_val = scaler.transform(X_val)
|
Multiclass Classification using LightGBM
Before training an actual lightGBM model using the dataset first we will have to convert the dataset into lgb Datasets using lgb.Dataset API.
Python3
params = {
'objective': 'multiclass',
'num_class': 4,
'metric': 'multi_logloss',
'verbose': 0
}
train_data = lgb.Dataset(X_train, label=Y_train)
valid_data = lgb.Dataset(X_val, label=Y_val, reference=train_data)
|
Now as the parameters and the dataset has been defined let’s train the model.
Python3
num_round = 100
model = lgb.train(params,
train_data,
num_round,
early_stopping_rounds=10,
valid_sets=[valid_data])
|
Output:
[80] valid_0's multi_logloss: 0.605381
[81] valid_0's multi_logloss: 0.60547
[82] valid_0's multi_logloss: 0.605018
[83] valid_0's multi_logloss: 0.605324
[84] valid_0's multi_logloss: 0.60554
[85] valid_0's multi_logloss: 0.605153
[86] valid_0's multi_logloss: 0.60477
Early stopping, best iteration is:
[76] valid_0's multi_logloss: 0.604612
Now we can use the trained model to create predictions on the validation data.
Python3
y_pred = model.predict(X_val,
num_iteration=model.best_iteration)
y_pred.shape
y_pred[:4]
|
Output:
array([[8.77103321e-01, 9.94841889e-02, 2.05788684e-02, 2.83362177e-03],
[1.19964248e-04, 1.08561258e-03, 1.06810300e-03, 9.97726320e-01],
[2.04789377e-04, 1.22397780e-03, 2.83356414e-02, 9.70235591e-01],
[6.00331842e-04, 2.70622699e-03, 9.55073045e-01, 4.16203966e-02]])
As we have got the probabilities for the four classes let’s use the argmax function to get the prediction classes.
Python3
y_pred = np.argmax(y_pred, axis=1)
y_pred[:10]
|
Output:
array([0, 3, 3, 2, 3, 0, 2, 3, 3, 2], dtype=int64)
Evaluation of the model
Now let’s evaluate the performance of the model using the validation data predictions.
Python3
accuracy = accuracy_score(Y_val, y_pred)
print(f'Accuracy: {accuracy}')
|
Output:
Accuracy: 0.7525195968645016
Let us check the classification report for the classes present in the dataset:
Python3
print(classification_report(Y_val, y_pred))
|
Output:
precision recall f1-score support
0 0.74 0.88 0.80 661
1 0.64 0.65 0.65 683
2 0.73 0.66 0.69 642
3 0.92 0.81 0.86 693
accuracy 0.75 2679
macro avg 0.76 0.75 0.75 2679
weighted avg 0.76 0.75 0.75 2679
Conclusion
LightGBM can be considered to be a powerful and efficient tool for multiclass classification tasks. It can be useful when we are in need of faster computation and to handle large datasets. This algorithm can leverage it’s abilities to provide accurate predictions in a time crunched period.
Share your thoughts in the comments
Please Login to comment...