Reading large numeric TSV file into memory in R
Last Updated :
20 Mar, 2024
TSV (Tab-Separated Values) files are a common way of storing data. When working with large numeric TSV files in the R programming language, memory issues can arise when reading them in. Fortunately, there are several R packages and functions available to help with this task, including readr and data.table. The readr package provides a read_tsv() function to read in delimited text files, while the data.table package offers tools for data manipulation and reading large files.
Reading large numeric TSV file
Working with large numeric TSV files can be a challenge when reading them into memory in R. These files are often too large to be loaded into memory at once, causing out-of-memory errors. However, there are several ways to address this issue, including using the data.table package and the fread() function. The fread() function is designed for the fast reading of large files in memory-efficient ways. It is particularly useful when dealing with large TSV files with a mix of data types.
Example of reading a large numeric TSV file into memory
Here’s an example of reading a large numeric TSV file into memory using the data.table package and providing sample code with outputs.
For this article, we will use the large_numeric_file.tsv file as an example.
Example 1: Reading large numeric TSV file into memory
We can use the following code to read the file into memory as a data table using data.table:
R
library(data.table)
DT <- fread("large_numeric_file.tsv")
head(DT)
|
Output:
In this example, we first load the data.table package and then use the fread() function to read the TSV file “large_numeric_file.tsv” into a data table called DT. The resulting data table DT contains the data read from the file, with one row per line and one column per tab-separated field in the file.
We then print the first few rows of the data table using the head() function to verify that the data has been read correctly.
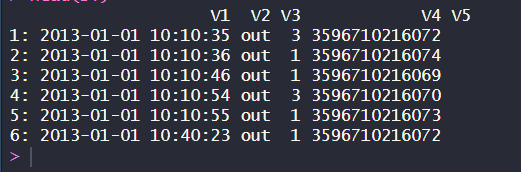
Reading large numeric TSV file
Example 2: Reading chunks of large numeric TSV file into memory
If the TSV file is too large to fit into memory, we can use the chunk argument of fread() to read the file in chunks. Here’s an example:
R
library(data.table)
chunk_size <- 3
for (chunk in seq(from = 1, to = nrow(fread("large_numeric_file.tsv",
nrows = 1)),
by = chunk_size)) {
DT <- fread("large_numeric_file.tsv", skip = chunk - 1,
nrows = chunk_size)
print(head(DT))
}
|
Output:
In this example, we set the chunk_size variable to 3, which means we will read the TSV file in chunks of 3 rows at a time. We then loop over the TSV file in chunks using the skip and nrows arguments of fread(). For each chunk, we create a new data table DT and do some processing on the chunk, which in this case is simply printing the first few rows of the data table.
As we can see from the output, the TSV file has been read correctly and processed in chunks.
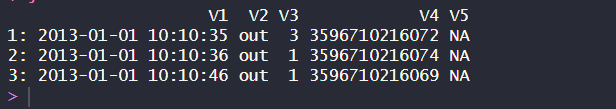
Reading chunks of large numeric TSV file
Example 3: Reading a few lines of a large numeric TSV file into memory
If we want to read some particular lines of ourTSV file into the memory, it can be done by slicing the dataframe in R.
R
library(data.table)
DT <- fread("large_numeric_file.tsv")
DT[3:7]
|
Output:
The above code will print the rows who has an index value of 3 to 7.
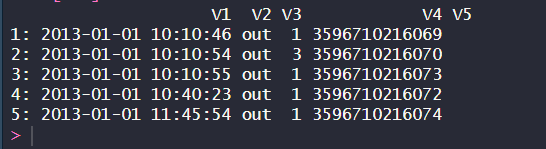
Reading a few lines of a large numeric TSV file
Share your thoughts in the comments
Please Login to comment...