Applications of String Matching Algorithms
Last Updated :
07 Nov, 2022
String matching algorithms have greatly influenced computer science and play an essential role in various real-world problems. It helps in performing time-efficient tasks in multiple domains. These algorithms are useful in the case of searching a string within another string. String matching is also used in the Database schema, Network systems.
Let us look at a few string matching algorithms before proceeding to their applications in real world. String Matching Algorithms can broadly be classified into two types of algorithms –
- Exact String Matching Algorithms
- Approximate String Matching Algorithms
Exact String Matching Algorithms:
Exact string matching algorithms is to find one, several, or all occurrences of a defined string (pattern) in a large string (text or sequences) such that each matching is perfect. All alphabets of patterns must be matched to corresponding matched subsequence. These are further classified into four categories:
- Algorithms based on character comparison:
- Naive Algorithm: It slides the pattern over text one by one and check for a match. If a match is found, then slides by 1 again to check for subsequent matches.
- KMP (Knuth Morris Pratt) Algorithm: The idea is whenever a mismatch is detected, we already know some of the characters in the text of the next window. So, we take advantage of this information to avoid matching the characters that we know will anyway match.
- Boyer Moore Algorithm: This algorithm uses best heuristics of Naive and KMP algorithm and starts matching from the last character of the pattern.
- Using the Trie data structure: It is used as an efficient information retrieval data structure. It stores the keys in form of a balanced BST.
- Deterministic Finite Automaton (DFA) method:
- Automaton Matcher Algorithm: It starts from the first state of the automata and the first character of the text. At every step, it considers next character of text, and look for the next state in the built finite automata and move to a new state.
- Algorithms based on Bit (parallelism method):
- Aho-Corasick Algorithm: It finds all words in O(n + m + z) time where n is the length of text and m be the total number characters in all words and z is total number of occurrences of words in text. This algorithm forms the basis of the original Unix command fgrep.
- Hashing-string matching algorithms:
- Rabin Karp Algorithm: It matches the hash value of the pattern with the hash value of current substring of text, and if the hash values match then only it starts matching individual characters.
Approximate String Matching Algorithms:
Approximate String Matching Algorithms (also known as Fuzzy String Searching) searches for substrings of the input string. More specifically, the approximate string matching approach is stated as follows: Suppose that we are given two strings, text T[1…n] and pattern P[1…m]. The task is to find all the occurrences of patterns in the text whose edit distance to the pattern is at most k. Some well known edit distances are – Levenshtein edit distance and Hamming edit distance.
These techniques are used when the quality of the text is low, there are spelling errors in the pattern or text, finding DNA subsequences after mutation, heterogeneous databases, etc. Some approximate string matching algorithms are:
- Naive Approach: It slides the pattern over text one by one and check for approximate matches. If they are found, then slides by 1 again to check for subsequent approximate matches.
- Sellers Algorithm (Dynamic Programming)
- Shift or Algorithm (Bitmap Algorithm)
Applications of String Matching Algorithms:
- Plagiarism Detection: The documents to be compared are decomposed into string tokens and compared using string matching algorithms. Thus, these algorithms are used to detect similarities between them and declare if the work is plagiarized or original.

- Bioinformatics and DNA Sequencing: Bioinformatics involves applying information technology and computer science to problems involving genetic sequences to find DNA patterns. String matching algorithms and DNA analysis are both collectively used for finding the occurrence of the pattern set.

- Digital Forensics: String matching algorithms are used to locate specific text strings of interest in the digital forensic text, which are useful for the investigation.
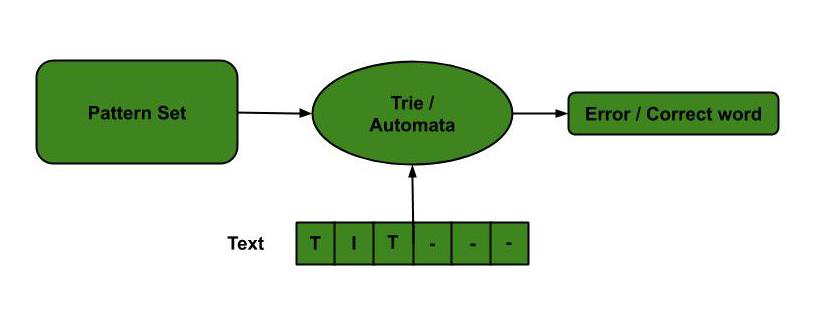
- Spelling Checker: Trie is built based on a predefined set of patterns. Then, this trie is used for string matching. The text is taken as input, and if any such pattern occurs, it is shown by reaching the acceptance state.
- Spam filters: Spam filters use string matching to discard the spam. For example, to categorize an email as spam or not, suspected spam keywords are searched in the content of the email by string matching algorithms. Hence, the content is classified as spam or not.

- Search engines or content search in large databases: To categorize and organize data efficiently, string matching algorithms are used. Categorization is done based on the search keywords. Thus, string matching algorithms make it easier for one to find the information they are searching for.

- Intrusion Detection System: The data packets containing intrusion-related keywords are found by applying string matching algorithms. All the malicious code is stored in the database, and every incoming data is compared with stored data. If a match is found, then the alarm is generated. It is based on exact string matching algorithms where each intruded packet must be detected.

Share your thoughts in the comments
Please Login to comment...