PCA and SVM Pipeline in Python
Last Updated :
04 Mar, 2024
Principal Component Analysis (PCA) and Support Vector Machines (SVM) are powerful techniques used in machine learning for dimensionality reduction and classification, respectively. Combining them into a pipeline can enhance the performance of the overall system, especially when dealing with high-dimensional data. The aim of the article is demonstrate how we can utilize PCA and SVM in single pipeline in Python.
What is a support vector machine?
Support Vector Machine is used for classification and regression problems. At first it was used for only classification problems. It works by finding the best hyperplane that separates the different classes. The hyperplane is selected by maximizing the margin between the classes.
What is PCA?
Principal Component Analysis is a technique used for dimensionality reduction in machine learning. It is useful for converting larger datasets into smaller datasets with maintaining all the patterns. It helps reduce the features in the data while preserving the maximum amount of information.
Why use Pipeline?
A machine learning pipeline is like an assembly line where many processes are are connected sequentially, such as preparing data, training the data, etc. So, it becomes easier to work with from start to end.
Even we can do our all our task from training to testing without the pipeline, so what is the real benefit of using pipeline? There are many reasons to use pipeline, let’s discuss few of them:
- When using pipeline, same preprocessing steps can be applied to both training and testing data without writing different code for both of them.
- It encapsulates the entire work into single object.
- It is most useful when label encoding has to be done for both training and testing.
- It reduces the code length as when we want to train for multiple times we can use that pipeline.
Implementation of PCA and SVM in a Pipeline
Importing Required Libraries
Python3
import pandas as pd
import seaborn as sns
from sklearn.svm import SVC
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
from sklearn.pipeline import Pipeline
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
|
Loading the dataset and Preprocessing
Python3
from sklearn import datasets
cancer = datasets.load_breast_cancer()
|
Convert loaded data into DataFrame
Python3
df = pd.DataFrame(data=cancer.data, columns=cancer.feature_names)
df['target'] = cancer.target
|
Understanding the data
Python3
print("Shape of the dataset:", df.shape)
print(df.target.unique())
df.sample(5)
|
Output:
Shape of the dataset: (569, 31)
[0 1]
mean radius mean texture mean perimeter mean area mean smoothness mean compactness mean concavity mean concave points mean symmetry mean fractal dimension ... worst texture worst perimeter worst area worst smoothness worst compactness worst concavity worst concave points worst symmetry worst fractal dimension target
86 14.48 21.46 94.25 648.2 0.09444 0.09947 0.12040 0.04938 0.2075 0.05636 ... 29.25 108.40 808.9 0.1306 0.1976 0.3349 0.12250 0.3020 0.06846 0
554 12.88 28.92 82.50 514.3 0.08123 0.05824 0.06195 0.02343 0.1566 0.05708 ... 35.74 88.84 595.7 0.1227 0.1620 0.2439 0.06493 0.2372 0.07242 1
221 13.56 13.90 88.59 561.3 0.10510 0.11920 0.07860 0.04451 0.1962 0.06303 ... 17.13 101.10 686.6 0.1376 0.2698 0.2577 0.09090 0.3065 0.08177 1
510 11.74 14.69 76.31 426.0 0.08099 0.09661 0.06726 0.02639 0.1499 0.06758 ... 17.60 81.25 473.8 0.1073 0.2793 0.2690 0.10560 0.2604 0.09879 1
118 15.78 22.91 105.70 782.6 0.11550 0.17520 0.21330 0.09479 0.2096 0.07331 ... 30.50 130.30 1272.0 0.1855 0.4925 0.7356 0.20340 0.3274 0.12520 0
5 rows × 31 columns
So as you can see the data consists of 5 rows and 31 columns, and in target column there are two classes 0 and 1, so it is a classification problem, so we will use Support Vector Machine, and as you can see there are 30 features so here we can apply principal component analysis for reducing the features.
Splitting Data in training and testing
Python3
X = df.drop(['target'], axis=1)
print("Shape of X:", X.shape)
y = df['target']
print("Shape of y:", y.shape)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=10)
print("Shape of X_train:", X_train.shape)
print("Shape of X_test:", X_test.shape)
print("Shape of y_train", y_train.shape)
print("Shape of y_test", y_test.shape)
|
Output:
Shape of X: (569, 30)
Shape of y: (569,)
Shape of X_train: (455, 30)
Shape of X_test: (114, 30)
Shape of y_train (455,)
Shape of y_test (114,)
Our data is spillted into 80% training and 20% testing and we have provided random state so if we rerun the code then also same spilliting will be done.
Creating Pipeline
Python3
clf = Pipeline([
("pca",PCA(n_components=0.7)),
("svm",SVC(C=10,kernel='rbf')),
])
clf.fit(X_train,y_train)
|
Output:
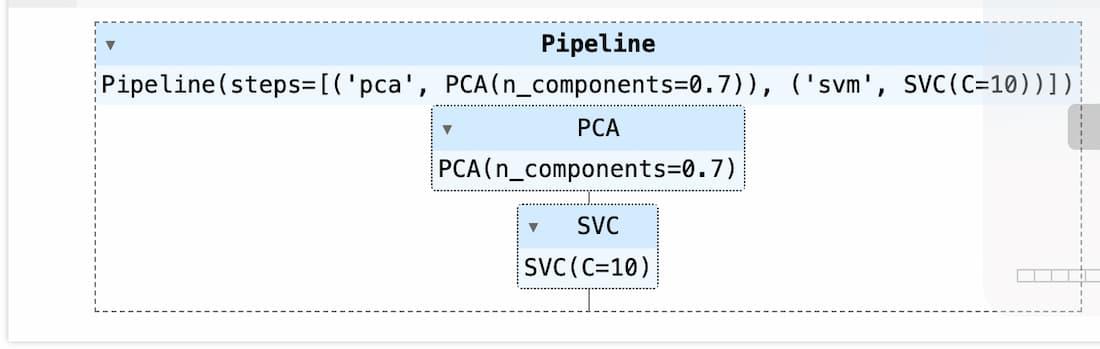
SVM with PCA
So, as you can see with pipeline, we don’t have to use any fit_transform instead it will be taken care by pipeline, additionally if there are more preprocessing steps you can include them in the pipeline and train it.
Using PCA and SVM in a pipeline streamlines the modeling process by combining preprocessing (dimensionality reduction) and modeling (SVM classification) into a single workflow. This simplifies code maintenance and facilitates reproducibility.
Prediction using the model
Python3
print("Score is:",clf.score(X_test,y_test))
y_pred = clf.predict(X_test)
report = classification_report(y_test, y_pred)
print("Classificaion Report")
print(report)
|
Output:
Score is: 0.8859649122807017
Classificaion Report
precision recall f1-score support
0 0.91 0.74 0.82 39
1 0.88 0.96 0.92 75
accuracy 0.89 114
macro avg 0.89 0.85 0.87 114
weighted avg 0.89 0.89 0.88 114
Confusion Matrix
Python3
cm = confusion_matrix(y_test,y_pred)
print(cm)
sns.heatmap(cm,annot=True)
plt.ylabel("Truth")
plt.xlabel("Predicted")
|
Output:
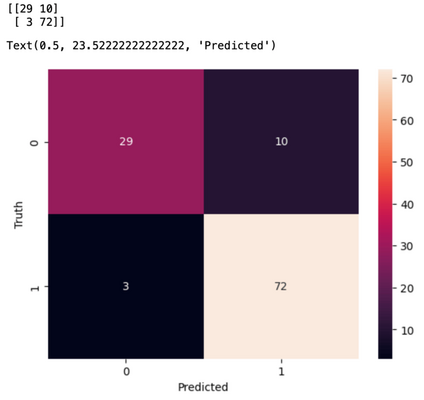
When we use Principal Component Analysis and Support Vector Machine together in pipeline, it becomes a smooth process for preparing our data before building our model. It is useful when data is complex and large and want to achieve a good accuracy on our model.
Share your thoughts in the comments
Please Login to comment...