Alzheimer’s detection Using Machine Learning
Last Updated :
31 Jul, 2023
Alzheimer’s disease is a neurodegenerative disorder characterized by progressive cognitive decline and memory loss. Detecting Alzheimer’s disease early is crucial for timely intervention and treatment. While I can provide some general information on Alzheimer’s detection, it’s important to consult medical professionals for accurate diagnosis and advice.
Alzheimer’s detection
Machine learning has shown promise in aiding the detection of Alzheimer’s disease. In this article, we will apply RandomForestClassifier and LocalOutlierFactor to detect Alzheimer’s disease.
Steps:
- Launch the Jupyter Notebook or Colab
- Download the dataset from the given link.
- Import required libraries.
- Preprocess the dataset.
- Perform Outlier detection.
- Train and Test model.
- Evaluate the model.
- You can use visualization techniques (option – if needed)
Download the dataset from here: Dataset
|
Group
|
Nondemented or Demented
|
|
EDUC
|
Years of education
|
|
SES
|
Socioeconomic Status
|
|
MMSE
|
Mini-Mental State Examination
|
|
CDR
|
Clinical Dementia Rating
|
|
eTIV
|
Estimated Total Intracranial Volume
|
|
nWBV
|
Normalize Whole Brain Volume
|
|
ASF
|
Atlas Scaling Factor
|
The dataset contains 373 rows with 14 fields which contain Group fields with classes Demented and Non-Demented where we need to work on them.
Import required libraries:
Python3
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import LocalOutlierFactor
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score, confusion_matrix
import seaborn as sns
import matplotlib.pyplot as plt
|
We have imported the required packages.
Load the dataset
Python3
df = pd.read_csv('oasis_longitudinal.csv')
print(df.head())
|
Output:
Subject ID MRI ID Group Visit MR Delay M/F Hand Age EDUC \
0 OAS2_0001 OAS2_0001_MR1 Nondemented 1 0 M R 87 14
1 OAS2_0001 OAS2_0001_MR2 Nondemented 2 457 M R 88 14
2 OAS2_0002 OAS2_0002_MR1 Demented 1 0 M R 75 12
3 OAS2_0002 OAS2_0002_MR2 Demented 2 560 M R 76 12
4 OAS2_0002 OAS2_0002_MR3 Demented 3 1895 M R 80 12
SES MMSE CDR eTIV nWBV ASF
0 2.0 27.0 0.0 1987 0.696 0.883
1 2.0 30.0 0.0 2004 0.681 0.876
2 NaN 23.0 0.5 1678 0.736 1.046
3 NaN 28.0 0.5 1738 0.713 1.010
4 NaN 22.0 0.5 1698 0.701 1.034
Check the data info
Output:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 373 entries, 0 to 372
Data columns (total 15 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Subject ID 373 non-null object
1 MRI ID 373 non-null object
2 Group 373 non-null object
3 Visit 373 non-null int64
4 MR Delay 373 non-null int64
5 M/F 373 non-null object
6 Hand 373 non-null object
7 Age 373 non-null int64
8 EDUC 373 non-null int64
9 SES 354 non-null float64
10 MMSE 371 non-null float64
11 CDR 373 non-null float64
12 eTIV 373 non-null int64
13 nWBV 373 non-null float64
14 ASF 373 non-null float64
dtypes: float64(5), int64(5), object(5)
memory usage: 43.8+ KB
Preprocessing the dataset
Dropping irrelevant columns
Python3
df = df.drop(['Subject ID', 'MRI ID', 'Hand', 'Visit', 'MR Delay'], axis=1)
df.shape
|
Output:
(373, 10)
Check Group Features
Python3
df['Group'].value_counts()
|
Output:
Group
Nondemented 190
Demented 146
Converted 37
Name: count, dtype: int64
Replace the “Converted” with “Demented”
Python3
df['Group'] = df['Group'].replace(['Converted'], ['Demented'])
|
Replace Categorical String Data with Numerical Data
- df[‘M/F’] = df[‘M/F’].replace([‘F’, ‘M’], [0, 1]): Replaces the values in the ‘M/F’ (Male/Female) column, ‘F’ with 0 and ‘M’ with 1. This helps to convert categorical values into binary format.
- df[‘Group’] = df[‘Group’].replace([‘Demented’, ‘Nondemented’], [1, 0]): Replaces the values ‘Demented’ with 1 and ‘Nondemented’ with 0 in the ‘Group’ column for binary classification.
Python3
df['M/F'] = df['M/F'].replace(['F', 'M'], [0, 1])
df['Group'] = df['Group'].replace(['Demented', 'Nondemented'], [1, 0])
|
Check for Null Values
Output:
Group 0
M/F 0
Age 0
EDUC 0
SES 19
MMSE 2
CDR 0
eTIV 0
nWBV 0
ASF 0
dtype: int64
Imputating the missing values
As we can see that only the SES column has null values. Because SES denotes ranks of Socioeconomic Status, It is a categorical feature. So, We can replace the null values of SES with its mode.
Fill the MMSE null values with its Median
Python3
df['SES'] = df['SES'].fillna(value= df['SES'].mode().iloc[0])
df['MMSE'] = df['MMSE'].fillna(value =df['MMSE'].median())
|
Separate Input Features and Target Variables Group
Python3
Y = df['Group'].values
X = df[['M/F', 'Age', 'EDUC', 'SES', 'MMSE', 'eTIV', 'nWBV', 'ASF']]
|
Y represents the target variable – labels as a Numpy array and X represents the list of selected columns as a matrix which is used for the classification task.
Numerical and Categorical Features
Python3
numerical_features = ['Age', 'EDUC', 'MMSE', 'eTIV', 'nWBV', 'ASF']
categorical_features = ['M/F', 'SES']
|
Treating Outliers
outlier_detector = LocalOutlierFactor(contamination=0.05): Creating an instance of the Local Outlier Factor (LOF) algorithm for outlier detection. The contamination parameter is set to 0.05, which means we are expecting 5% outliers.
outliers = outlier_detector.fit_predict(X): This step calculates the outlier scores for each sample. This also gives and assigns a label of -1 for outliers and 1 for inliers.
Python3
outlier_detector = LocalOutlierFactor(contamination=0.05)
outliers = outlier_detector.fit_predict(X)
X = X[outliers == 1]
Y = Y[outliers == 1]
|
Standardize and One Hot Encoding
The next two steps represent that we are selecting only inliers for further processing. This step of code creates transformers for categorical numerical and numerical features and helps in training the model consistently.
- MinMaxScalar helps to scale numerical features.
- OneHotEncoder is used to encode categorical features.
- These transformers are merged using a ColumnTransformer.
Python3
numerical = MinMaxScaler()
categorical = OneHotEncoder(drop='first')
p = ColumnTransformer(transformers=[('num', numerical, numerical_features),
('cat', categorical, categorical_features)])
|
Visualizing dataset target class details:
Python3
sns.countplot(data=df, x='Group')
plt.title("Distribution of Classes")
plt.xlabel("Group")
plt.ylabel("Count")
plt.show()
|
Output:
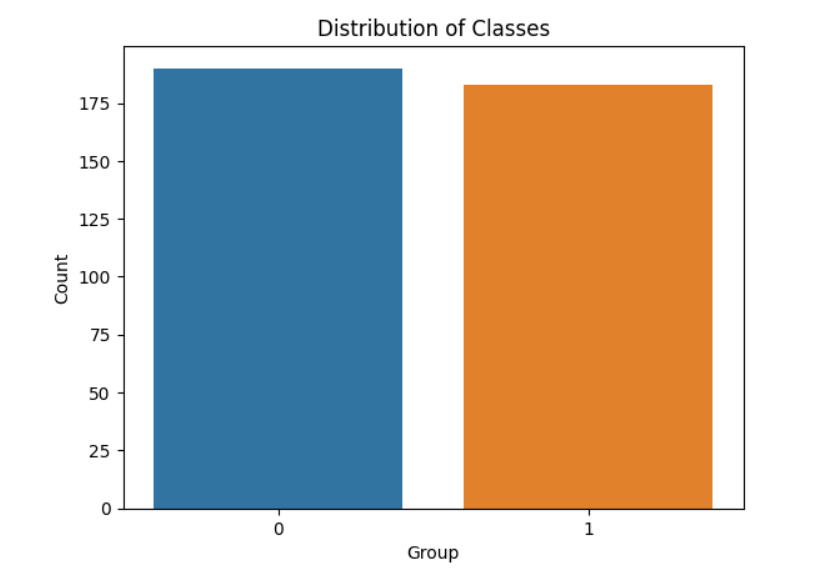
Target class distribution
Splitting data for training and testing:
Python3
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, random_state=0)
|
By default, 75% for training and 25% for testing.
Build the Pipeline
Building a pipeline for usage with Random Forest classifier and Preprocessor, then fit the model.
Python3
pl_rf = Pipeline(steps=[('preprocessor', p),('classifier', RandomForestClassifier(random_state=0))])
pl_rf.fit(X_train, Y_train)
|
Predictions:
Python3
Y_pred_rf = pl_rf.predict(X_test)
|
Evaluating the model:
Python3
print("Accuracy:", accuracy_score(Y_test, Y_pred_rf))
print("Precision:", precision_score(Y_test, Y_pred_rf))
print("Recall:", recall_score(Y_test, Y_pred_rf))
print("F1 Score:", f1_score(Y_test, Y_pred_rf))
|
Output:
Accuracy: 0.8651685393258427
Precision: 0.825
Recall: 0.868421052631579
F1 Score: 0.8461538461538461
Confusion Matrix:
Python3
cnf = confusion_matrix(Y_test, Y_pred_rf)
sns.heatmap(cnf, annot=True, fmt='d', cmap='Green')
plt.title("Confusion Matrix for above model")
plt.xlabel("Predicted")
plt.ylabel("True")
plt.show()
|
Output:
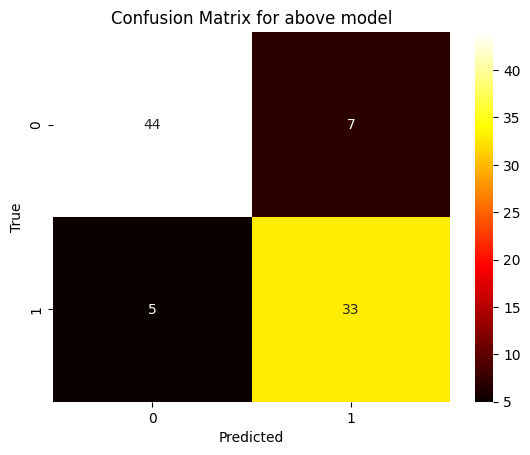
Confusion Matrix
Share your thoughts in the comments
Please Login to comment...