The scipy.stats is the SciPy sub-package. It is mainly used for probabilistic distributions and statistical operations. There is a wide range of probability functions.
There are three classes:
Class
| Description
|
| rv_continuous | For continuous random variables, we can create specialized distribution subclasses and instances. |
| rv_discrete | For discrete random variables, we can create specialized distribution subclasses and instances. |
| rv_histogram | generate specific distribution histograms. |
Continuous Random Variables
A continuous random variable is a probability distribution when the random variable X can have any value. The mean is defined by the location (loc) keyword. The standard deviation is determined by the scale (scale) keyword.
As we discussed that using the rv_continuous class we can create distributed subclasses and instances so there is a method called ‘norm’ which inherits from rv_continuous and this function will calculate the CDF for us.
Let X be a continuous random variable with PDF( (f) and CDF (F).
PDF – Probability Density Function
The PDF of a continuous random variable x satisfies the following conditions. If f\left ( x \right )\geq 0 for all x\in \mathbb{R} here f is piecewise continuous.
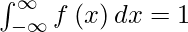
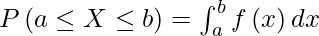
The CDF is found by integrating the PDF:
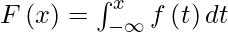
The pdf can be found by differentiating the CDF:
![Rendered by QuickLaTeX.com f\left ( x \right )=\frac{\mathrm{d} }{\mathrm{d} x}\left [ F\left ( x \right ) \right ]](https://www.geeksforgeeks.org/wp-content/ql-cache/quicklatex.com-1d8f14ce28c48f1789b052b5571108e9_l3.png)
Python3
import numpy as npy
from scipy.stats import norm
print(norm.cdf(npy.array([-2, 0, 2])))
|
Output:
[0.02275013 0.5 0.97724987]
Discrete Random Variables
Only a countable number of values can be assigned to discrete random variables. L is an additional integer parameter that can be added to any discrete distribution. The general distribution p and the standard distribution p0 have the following relationship:
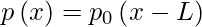
scipy.stats.circmean
Compute the circular mean for samples in a range. We will use the following function to calculate the circular mean:
Syntax:
scipy.stats.circmean(array, high=2*pi, low=0, axis=None, nan_policy=’propagate’)
where,
- Array – input array or samples.
- high (float or int ) – high boundary for sample. default high = 2 * pi.
- low ( float or int ) – low boundary for sample. default low = 0.
- axis ( int ) – Axis along which means are computed.
- nan_policy ( ‘propagate’, ‘raise’, ‘omit’ ) – Defines how to handle when input contains nan. ‘propagate’ returns nan, ‘raise’ throws an error, and ‘omit’ performs the calculations ignoring nan values. The default is ‘propagate’.
Python3
from scipy.stats import circmean
print(circmean([0.4, 2.4, 3.6], high=4, low=2))
|
Output:
2.254068341376122
scipy.stats.contingency.crosstab
Given the lists a and p, create a contingency table that counts the frequencies of the corresponding pairs.
Python3
from scipy.stats.contingency import crosstab
a = ['A', 'B', 'A', 'A', 'B', 'B', 'A', 'A', 'B', 'B']
p = ['P', 'P', 'P', 'Q', 'R', 'R', 'Q', 'Q', 'R', 'R']
print(crosstab(a, p))
(auv, puv), cnt = crosstab(a, p)
print(auv)
print(puv)
print(cnt)
|
Output:
((array(['A', 'B'], dtype='<U1'), array(['P', 'Q', 'R'], dtype='<U1')), array([[2, 3, 0],
[1, 0, 4]]))
['A' 'B']
['P' 'Q' 'R']
[[2 3 0]
[1 0 4]]
Note – In the above output, we have a ndarray, which consists of the different other arrays. The first value (array([‘A’, ‘B’]), dtype='<U1′) is basically the array of unique values in the list a, the second value (array([‘P’, ‘Q’, ‘R’]),dtype='<U1′) is basically the array of unique values in the list p, and the third value is the frequency of each pair of list a and list p.
list a =
list b =
Result analysis
Above image observations –
A - P = 2
A - Q = 3
A - R = 0
Above image observations:
B - P = 1
B - Q = 0
B - R = 4
stats.describe()
This function basically calculates the several descriptive statistics of the argument array.
Syntax:
scipy.stats.describe(a, axis=0, ddof=1, bias=True, nan_policy=’propagate’)
where,
- Input array – array for which we want to generate the statistics.
- axis ( int , float ) { # optional } – Axis along which statistics are calculated. The default axis is 0.
- ddof ( int ) { # optional } – Delta Degrees for variance. Default ddof = 1.
- bias ( bool ) { # optional } – skewness and kurtosis calculations for statistical bias.
- nan_policy – { ‘propagate’,’raise’,’omit’ } { # optional ) – Handle the NAN inputs.
Return:
- nbos ( int or ndarray ) – length of data along axis value.
- minmax ( tuple of ndarrays or floats ) – Minimum and Maximum value of input array along the given axis.
- mean ( float or ndarray ) – mean of input array.
- variance ( ndarray or float ) – variance of input array along the given axis.
- skewness ( float or ndarray ) – skewness of input array along the given axis.
- kurtosis ( ndarray or float ) – kurtosis of input array along the given axis.
Python3
from scipy import stats as st
import numpy as npy
array = npy.array([10, 20, 30, 40, 50, 60, 70, 80])
print(st.describe(array))
|
Output:
DescribeResult(
nobs=8,
minmax=(10, 80),
mean=45.0,
variance=600.0,
skewness=0.0,
kurtosis=-1.2380952380952381)
Python3
from scipy import stats as st
import numpy as npy
nd = npy.array([[5, 6], [2, 3], [5, 5],\
[7, 9], [9, 8], [8, 7]])
print(st.describe(nd))
|
Output:
DescribeResult(nobs=6,
minmax=(array([2, 3]),
array([9, 9])),
mean=array([6. , 6.33333333]),
variance=array([6.4 , 4.66666667]),
skewness=array([-0.40594941, -0.3380617 ]),
kurtosis=array([-0.9140625, -0.96 ]))
scipy.stats.kurtosis
Kurtosis quantifies how much of a probability distribution’s data are concentrated towards the mean as opposed to the tails.
Kurtosis is the fourth central moment divided by the square of the variance.
Syntax:
scipy.stats.kurtosis(a, axis=0, fisher=True, bias=True, nan_policy=’propagate’, *, keepdims=False
where,
- Input array – Data for which the kurtosis is calculated..
- axis ( int , float ) { # optional } – Axis along which statistics are calculated. The default axis is 0.
- fisher ( bool ) { # optional } – If True, Fisher’s definition is used. If False, Pearson’s definition is used.
- bias ( bool ) { # optional } – If False, then the calculations are corrected for statistical bias.
- nan_policy – { ‘propagate’,’raise’,’omit’ } { # optional ) – Handle the NAN inputs.
- keepdims( bool ) ( # optional ) – default is false. broadcast result correctly against the input array.
Returns:
- kurtosis array – along the given axis.
Python3
from scipy import stats as st
dataset = st.norm.rvs(size=88)
print(st.kurtosis(dataset))
|
Output:
0.04606780907050423
scipy.stats.mstats.zscore
The Z-score provides information on how far a given value deviates from the standard deviation. When a data point’s Z-score is 0, it means that it has the same score as the mean.
Z = ( Observed Value ( x ) – mean ( μ ) ) / standard deviation ( σ )
Calculate the z score for each value in the input array in comparison to the sample mean and standard deviation.
Function parameters –
Syntax:
scipy.stats.mstats.zscore(a, axis=0, ddof=0, nan_policy=’propagate’)
where,
- Input array – sample input array.
- axis ( int , float ) { # optional } – Axis along which statistics are calculated. The default axis is 0.
- ddof ( int ) { # optional } – Degrees of freedom correction in the calculation of the standard deviation. The default value of ddof is 0.
- nan_policy – { ‘propagate’,’raise’,’omit’ } { # optional ) – Handle the NAN inputs.
Returns:
- zscore – array – The z-scores of input array a, normalised by mean and standard deviation.
Python3
from scipy import stats as st
dataset = [0.02, 0.5, 0.01, 0.33, 0.51, 1.0, 0.03]
nd = [[5.1, 6.1], [2.1, 3.1], [5.1, 5.1],\
[7.1, 9.1], [9.1, 8.1], [8.1, 7.1]]
print(st.zscore(dataset))
print(st.zscore(nd))
|
Output:
[-0.95649434 0.46555034 -0.98612027 -0.03809048 0.49517627 1.94684689
-0.92686841]
[[-0.4330127 -0.16903085]
[-1.73205081 -1.69030851]
[-0.4330127 -0.6761234 ]
[ 0.4330127 1.35224681]
[ 1.29903811 0.84515425]
[ 0.8660254 0.3380617 ]]
scipy.stats.skew
We can determine the direction of outliers from skewness. The tail of a distribution curve has a longer right side when there is a positive skew. Accordingly, the distribution curve’s outliers are farther from the mean on the left and closer to it on the right. Skewness just conveys the direction of outliers; it doesn’t provide information on the number of outliers.
Compute the sample skewness of a data set. Skewness should be close to zero for normally distributed data. A skewness value greater than zero indicates that the right tail of a unimodal continuous distribution has more weight.
Syntax:
scipy.stats.skew(a, axis=0, bias=True, nan_policy=’propagate’, *, keepdims=False)
where,
- Input array
- axis ( int , float ) { # optional } – Axis along which statistics are calculated. The default axis is 0.
- bias ( bool ) { # optional } – If False, then the calculations are corrected for statistical bias.
- nan_policy – { ‘propagate’,’raise’,’omit’ } { # optional ) – Handle the NAN inputs.
- keepdims( bool ) ( # optional ) – default is false. broadcast result correctly against the input array.
Return:
Python3
from scipy import stats as st
array = [99, 10, 30, 55, 50, 0, 90, 0]
print(st.skew(array))
|
Output:
0.3260023450293658
scipy.stats.energy_distance
Distance between two probability distributions. Suppose two distributions u and v and their CDF are U and V, two random variables X and Y are there, then the energy distance will be the square root of:
D2(U,V) = 2E || X – Y || – E || X – X’ || – E || Y – Y’ || > 0,
- || denotes the length of a vector
Compute the energy distance between two 1D distributions.
Python3
from scipy import stats as st
print(st.energy_distance([5, 10], [10, 20],\
[20, 30], [30, 40]))
|
Output:
2.851422845685634
scipy.stats.mode
Return an array of the most common values in the input array.
Python3
from scipy import stats as st
array = [[2, 3], [3, 1], [1, 3],\
[3, 3], [4, 2], [4, 4],\
[1, 2], [5, 6]]
print(st.mode(array))
|
Output:
ModeResult(mode=array([[1, 3]]), count=array([[2, 3]]))
scipy.stats.variation
The coefficient of variation – Standard deviation divided by the mean.
Python3
from scipy import stats as st
array = [[2, 3], [3, 1], [1, 3],\
[3, 3], [4, 2], [4, 4],\
[1, 2], [5, 6]]
print(st.variation(array, ddof=1))
|
Output:
[0.5070393 0.50395263]
scipy.stats.rankdata
Assign ranks to data, dealing with ties appropriately.
Python3
from scipy import stats as st
array = [2, 3, 15, 1, 6, 9, 8, 4, 5, 10]
print(st.rankdata(array))
|
Output:
[ 2. 3. 10. 1. 6. 8. 7. 4. 5. 9.]
Like Article
Suggest improvement
Share your thoughts in the comments
Please Login to comment...