CBSE Class 12 Molecular Basis of Inheritance: Inheritance is transmitted by certain molecules that Mendel termed as ‘factors’, but their nature was discovered later with the development of various scientific techniques. The molecules which govern the inheritance are called genes and it is of two types; Deoxyribonucleic acid (DNA) and Ribonucleic acid (RNA). In all organisms except viruses, DNA acts as the genetic material whereas RNA acts as the messenger molecule, however in some viruses RNA functions as the genetic material. It is because DNA is a more stable molecule as compared to RNA and hence, it evolved as the genetic material of almost all the organisms.
Search for Genetic Material
Many scientists performed multiple experiments that gave evidence that DNA is the genetic material. We will discuss the following three;
Transforming Principle
It was proposed by Frederick Griffith in 1928. He experimented on the pneumonia-causing bacteria Streptococcus pneumoniae to perform a bacterial transformation (transfer of genetic material from one bacterium to another). There are two strains of pneumococcus bacteria;
S-strain (Smooth strain)
|
R-strain (Rough strain)
|
| The virulent strain that causes infection |
The non-virulent strain |
| Have a polysaccharide coat |
Lacks polysaccharide coat |
| Produces smooth colonies |
Produces rough colonies |
Griffith’s Experiment includes the following steps;
- He injected live S-strain into mice –> Mice dies due to infection
- He injected live R-strain into mice –> Mice lives
- He injected heat-killed S-strain into mice –> Mice lives
- He injected a combination of heat-killed S-strain and live R-strain into the mice –> Mice die due to infection.
From the above experiment, he concluded that the R-strain bacteria had somehow been transformed by the heat-killed S-strain bacteria due to the transfer of a certain ‘transforming substance’ possibly the genetic material.
Biochemical Characterisation of Transforming Principle
It was performed by Oswald Avery, Colin MacLeod, and Maclyn McCarty (1944). They worked to determine the biochemical nature of the ‘transforming principle’ in Griffith’s experiment. they concluded that only DNase destroyed the transforming substance and not protease for protein and RNAase for RNA. Hence, the transforming substance is DNA and it is the genetic material.
Experiment to confirm DNA as the genetic material
Alfred Hershey and Martha Chase in 1952 proved that DNA is the genetic material. They used bacteriophages, radioactive phosphorus (P-32), and radioactive sulfur (S-35) to label them. They found that E.coli which were infected with viruses that had radioactive DNA were radioactive whereas bacteria that were infected with viruses that had radioactive proteins were not radioactive. It proved that when the virus infects a bacterium, only viral DNA gets into the bacterium and the viral protein remains outside.
The DNA
It is an acidic long polymer of deoxyribonucleotide that forms two complementary strands that run antiparallel to each other and are held together by hydrogen bonds between their opposite nitrogenous bases. It is made up of nucleotides which make the length of the DNA. Some of the examples are;
- Human = 6.6×109 BP,
- E.Coli =4.6×106 BP,
- Lambda phage =48502 BP, Ø X 174 phage =5386 nucleotides (Single-stranded DNA).
Structure of the polynucleotide chain
A polynucleotide is a polymer of nucleotides. Both DNA & RNA are polynucleotides. It is made up of three components; a nitrogenous base, a pentose sugar, and a phosphate group.
The nitrogenous bases are of two types; purines [Adenine (A) and Guanine (G)], and pyrimidines [Cytosine (C), Thymine (T) & Uracil (U)]. Note that thymine is only present in DNA and in the case of RNA Uracil is present instead of thymine. Pentose sugar is ribose in RNA and deoxyribose in DNA. And the phosphate group makes the nucleotide acidic. The nitrogenous base is linked to the pentose sugar through an N-glycosidic linkage to form nucleosides. The phosphate links two nucleotides to form a dinucleotide with the help of phosphodiester bonds.
- Nitrogenous base + sugar = Nucleoside
- Nucleoside + phosphate group = Nucleotide
Each polynucleotide chain has 2 free ends- a 3 prime (3’) end and the opposite 5 prime (5’) end. In the 3’ end, the 3rd C-atom of the sugar is free, i.e., it is not linked to any nucleotide. Similarly, in the 5’ end, the 5th C of sugar is free.
Structure of DNA
Based on the X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin, James Watson, and Francis Crick proposed a very simple but famous double helix model of DNA in 1953 which was seconded by the finding of Erwin Chargaff for a double-stranded DNA. The findings were;
- The purines and pyrimidines are always equal in amount i.e. A+G = T+C or A+G/T+C=1.
- The amount of adenine will always be equal to thymine and the amount of guanine will always be equal to cytosine i.e. A=T and G=C.
- Adenine is joined to thymine with 2 H-bonds while guanine is joined to cytosine by 3 H-bond.
Salient features of the Double Helical Structure of the DNA are;
- DNA is made of two polynucleotide chains, where the backbone is made of sugar-phosphate, and the bases are projected towards the inside.
- The two strands have anti-parallel polarity. It means if one strand has the polarity 5′ –> 3′, then the other has 3′ –> 5′.
- The bases in two strands are paired through hydrogen bonds (H-bonds) forming base pairs (bp). Purine always comes opposite to pyrimidine. This generates approximately uniform distance between the two strands of the helix (20Å).
- The double-stranded structure is coiled in a right-handed fashion. The pitch of the helix is 3.4 nm, and there is roughly 10 bp in each turn which makes the distance between a bp in a helix approximately equal to 0.34 nm.
- The plane of one base pair stacks over the other in a double helix pattern along with the H-bonds conferring stability of the helical structure.
Packaging of the DNA helix
The length of DNA is far greater than the dimension of a typical nucleus i.e. around 2.2 meters. This long-sized DNA is compacted and packed in the relatively smaller region of the nucleus.
In Prokaryotes the DNA is not packed inside a nucleus but it is compacted to form a structure called the nucleoid. In the nucleoid region, the negatively charged DNA is held by some positively charged non-histone proteins forming a looped structure.
In Eukaryotes the DNA is packed inside a membranous structure called the nucleus. Inside the nucleus, the DNA is wrapped around a unit of 8 molecules of positively charged protein histone in the form of histone octamer to form a structure called nucleosome that contains around 200 bp of DNA helix. The nucleosomes in turn form repeating units like “beads-on-string” structures in the nucleus called chromatin. Chromatin is packaged to form chromatin fibers which are further coiled and condensed at the metaphase stage of cell division to form chromosomes.
The RNA
It is the first genetic material that came into the earth. All life’s important metabolic processes developed around RNA. It is formed of a single polynucleotide chain that acts as the genetic material in some viruses called retroviruses like HIV. It acts as a genetic material as well as a catalyst enzyme like ribozyme. RNA being a catalyst was reactive and hence unstable. Therefore, DNA has evolved from RNA with chemical modifications that make it more stable.
DNA vs RNA
|
DNA
|
RNA
|
| Double-stranded |
Usually single-stranded |
| Contains deoxyribose sugar |
Contains ribose sugar |
| Have A, T, G, and C as nitrogenous bases |
Have A, U, G, and C as nitrogenous bases |
| Present in the nucleus, mitochondria, and chloroplast only. |
Present in cytoplasm, ribosome, and nucleolus. |
| It is stable as it is less reactive |
It is unstable as it is more reactive |
For more information read: DNA vs RNA
Properties of genetic material
- It should be able to generate its replica.
- It should be chemically and structurally stable.
- It should provide scope for slow changes (mutation) that are required for evolution.
- It should be able to express itself in the form of ‘Mendelian characters’.
Central Dogma
It is the pathway to how genetic information is converted into functional proteins that form the basis of living organisms. It was proposed by Francis Crick.
The flow is; DNA –> RNA –> Protein
However, in RNA viruses this flow of information takes place in the reverse direction RNA –> DNA –> RNA –> Protein
DNA Replication
The process of copying and making an identical copy of a double-stranded DNA from the parental DNA. In 1953 Watson & Crick proposed a Semi-conservative model of replication. It suggests that the 2 parental DNA strands separate and each act as the template for the synthesis of new complementary strands. After the completion of replication, each DNA molecule would have one parental and one new strand. However, the experimental proof was given by two different groups of scientists;
- Messelson and Stahl’s (1958) experiment on E. coli. They cultured the bacterium in a medium containing 15N (a heavy isotope of N) as the only nitrogen source for many generations that incorporated the 15N into DNA, making it heavier, and then they transferred the cells to a medium containing normal 14N, extracted the DNA, centrifuged it with CsCl, and measured their densities. This led to the conclusion that the newly synthesized DNA obtains one of its strands from the parent i.e. the replication is semi-conservative.
- Taylor and colleagues (1958) experimented on Vicia faba (fava beans). They performed a similar experiment on fava beans with radioactive thymidine to detect the distribution of the newly synthesized DNA in the chromosomes. They concluded that DNA in chromosomes also replicates semi-conservatively.
Mechanism of DNA Replication
In eukaryotes, DNA replication takes place at the S phase of the cell cycle. The process starts at the ORI point or Origin of Replication. The enzyme helicase will then help in unwinding the DNA by breaking the hydrogen bonds that separate the strand to make them the template strands for the new strand formation. These separated strands are stabilized by single-stranded binding proteins. During unwinding the tension in the strands is released by the enzyme topoisomerase. As this process is energetically expensive that is why during replication 2 strands of DNA cannot be separated in its entire length. This results in the formation of a Y-shaped replication fork. A small strand of RNA called RNA primer is synthesized at the 5’ end of the new DNA strand which is required for the initiation of replication by the DNA polymerase enzyme from the 5’ –> 3’ direction. The DNA polymerase enzyme forms one new strand (leading strand) in a continuous stretch in a 5’ –> 3’ direction. The other new strand is formed in a small fragment called the Okazaki fragment (lagging strand) which is discontinuous in the 3’ –> 5’ direction. These Okazaki fragments are joined together to form a new strand by DNA ligase.

DNA Transcription
It is the process of copying genetic information from one strand of DNA into RNA. In transcription, only a segment of DNA and only one of the two strands is copied into RNA. The enzyme involved in transcription is DNA-dependent RNA polymerase. The strand with polarity 3’ –> 5’ acts as a template for mRNA synthesis, and the other strand with polarity 5’ –> 3’ is called the coding strand (It does not code for anything). The transcriptional unit comprises a promoter, the structural gene, and a terminator. The promoter is the site where DNA-dependent RNA polymerase binds and is located in the 5’ end. The mRNA is produced from the structural gene. Terminator is the site at which transcription stops and is located in the 3’ end.
Mechanism of Transcription
The process of transcription is the same in both prokaryotes and eukaryotes. It consists of 3 steps;
- Initiation: Transcription is initiated at the promoter region. The sigma factor (σ factor/initiation factor) binds with RNA polymerase that binds in the promoter region.
- Elongation: The RNA chain is synthesized in a 5’ –> 3’ direction by the RNA polymerase enzyme by adding the nucleotides to the growing chain.
- Termination: Once the RNA polymerase reaches the terminator region of DNA, the rho factor (termination factor/ρ factor) binds to it and terminates the transcription.
However, in eukaryotes there are two additional complexities are present;
- There are 3 RNA polymerase enzymes present i.e. RNA Pol I, RNA Pol II, and RNA Pol III.
- The primary transcripts (hnRNA) contain both exons and introns and are non-functional. Hence introns have to be removed. For this, it undergoes firstly capping (a nucleotide methyl guanosine triphosphate cap is added to the 5’ end), then splicing (introns or non-coding regions are removed and exons or coding regions are spliced/joined together), and lastly tailing (200-300 adenylate residue are added at the 3’ end). Only after the end of these three steps, the hnRNA will be called mRNA and these three steps are collectively called post-transcriptional modifications.
In a cell there are three types of RNA found;
- mRNA (messenger RNA): It provides the template for translation or protein synthesis.
- rRNA (ribosomal RNA): It is the structural & catalytic RNA that functions during translation. E.g. ribozyme.
- tRNA (transfer RNA or sRNA or soluble RNA, being smallest): It is also called the adaptor molecule that carries specific amino acids for protein synthesis and reads the genetic code. It has an anticodon loop to read the codons and an amino acid acceptor end to which amino acids bind.
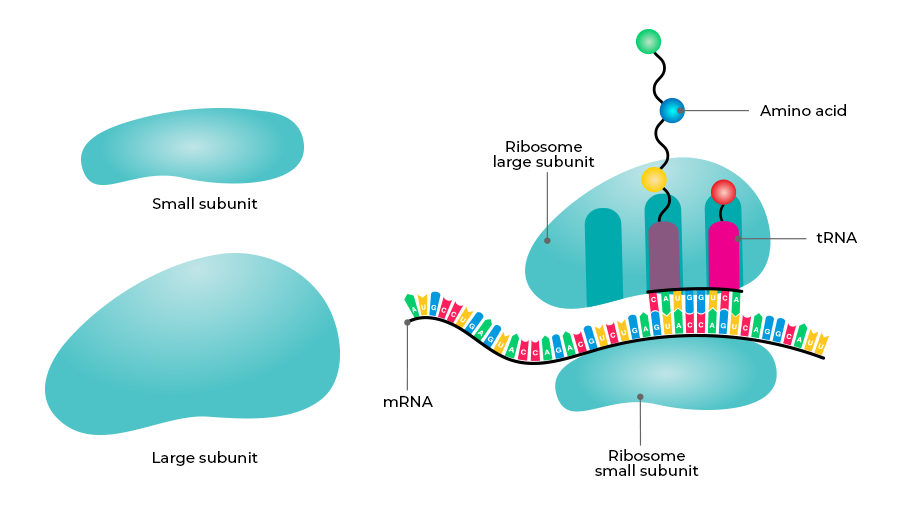
Translation
DNA Translation is the process in which the polymerization of amino acids to form a polypeptide chain or protein takes place. The order and sequence of amino acids in a protein is defined by the sequence of triplet codon in the mRNA. In the formed Protein, amino acids are joined by peptide bonds. The process of translation consists of 4 steps;
- Charging of tRNA: It is also called aminoacylation of tRNA. Amino acids are linked to their tRNA in the presence of aminoacyl tRNA synthetase enzyme and ATP. So the tRNA becomes charged.
- Initiation: It begins at the 5’ end of the mRNA where the small subunit of ribosome binds to mRNA. The initiator tRNA (methionyl tRNA) having UAC at the anticodon site blinds to the initiation codon on the mRNA. The large subunit of ribosome then blinds to mRNA that has two binding sites for tRNA – A site and P site. The initiator tRNA is found at the P site.
- Elongation: The other tRNA first binds to the A site and then shifts to the P site. Methionine from the first tRNA is now attached to the amino acid of the second tRNA through a peptide bond. The first tRNA is removed from the P site and the second tRNA is moved to the P site from the A site (translocation). Then again the third tRNA with amino acid binds at the A site. This process of peptide bond formation and translocation is repeated.
- Termination: When aminoacyl tRNA reaches the termination codon like UAA, UAG & UGA, the termination of translation occurs. The polypeptide and tRNA are released from the ribosomes and the ribosome dissociates into large and small subunits at the end of protein synthesis.
Regulation of Gene Expression
Gene expression results in the formation of a polypeptide that is required to perform a particular function or set of functions. Expression of the gene should be at the right time, at the correct amount. Otherwise, energy deprivation and metabolic chaos would occur in the cell. It can be regulated at several levels. In eukaryotes, the regulation could be exerted at;
- Transcriptional level (Formation of primary transcript)
- Processing level (Regulation of splicing)
- Transport of mRNA from the nucleus to the cytoplasm.
- Translational level
In prokaryotes, control of the rate of transcriptional initiation is the predominant site for control of gene expression. The set of genes regulating a metabolic reaction constitutes an Operon e.g. lac operon, trp operon, etc. When a substrate is added to the growth medium of bacteria, a set of genes is switched on to metabolize it. This is called induction. When a metabolite (product) is added, the genes to produce it are turned off. This is called repression.
Lac operon
It was proposed by Francois Jacob and Jacque Monod. The operon controlling lactose metabolism is called Lac Operon. It consists of;
- Three structural genes: Lac z gene that codes for Beta-galactosidase (It hydrolyzes lactose to glucose and galactose), Lac y gene that codes for Permease enzyme (Increases permeability of the cell to Lactose), and Lac a gene that codes for Transacetylase.
- A regulator gene (i gene/ inhibitor gene): It codes for a repressor protein.
- Inducer: Here lactose is the inducer that keeps the switch on and allows the structural gene to transcribe mRNA to synthesize the enzymes.
Functioning of Lac operon;
- When lactose (inducer) is absent: The regulator gene synthesizes mRNA and produces repressor protein which then binds to the operator region (blocks RNA polymerase movement). This prevents the transcription of mRNA from the structural gene (remains switched off).
- When lactose (inducer) is present: Lactose binds to the repressor protein making it inactive and it fails to bind to the operator region. The operator gene becomes free and induces the RNA polymerase to bind with the promoter gene (lac operon “switched on”) and transcribes structural genes.
Genetic Code
Genetic Code is the sequence of nucleotides (Nitrogen bases) in the mRNA that contains information for Protein Synthesis (Translation). The code is made up of 3 nitrogen bases or codons. This is called triplet code. There are 64 codons for 20 naturally occurring amino acids. Some of the significant scientists in this field were;
Salient Features of the Genetic Code
- The codon is Triplet i.e. there are 61 codons coding for amino acids and 3 codons are stop codons.
- The code is degenerate i.e. some amino acids are coded by more than one codon.
- One codon codes for only one amino acid, hence, it is unambiguous and specific.
- The codon is read on mRNA in a contiguous fashion i.e., there are no punctuations.
- The code is nearly universal. (eg: UUU codes for phenylalanine from bacteria to humans).
- AUG has a dual function. It codes for methionine and acts as an initiator codon/start codon.
- UAA, UAG, and UGA- Stop terminator codons.
Human Genome Project (HGP)
It was started in the year 1990 to map the entire human genome and completed in 2003. It was coordinated by the U.S. Department of Energy and the National Institute of Health along with many other research organizations in the world.
Its goals were;
- To identify all the genes in human DNA.
- To determine the sequence of 3 billion base pairs in human DNA.
- To store this information in databases.
- To address ethical, legal, and social issues (ELSI) that may arise from the project.
The methodologies of HGP are;
- Expressed sequence tags (ESTs): Focussed on identifying all genes that are expressed as RNA.
- Sequence annotation: Blind approach of sequencing the whole genome containing coding and non-coding sequences, needing vectors like BAC (Bacterial artificial chromosomes) and YAC (Yeast Artificial Chromosomes).
Process of HGP;
- The whole DNA from a cell is isolated
- Then it is converted into random fragments of relatively smaller sizes with the help of restriction enzymes.
- Those fragments are cloned in a suitable host (e.g.: bacteria and yeast) using specialized vectors (BAC and YAC) for amplification (now by PCR).
- The fragments are then sequenced using automated DNA sequencers (using the Frederick-Sanger method).
- These sequences were then arranged based on some overlapping regions present in them.
- Then all the sequences were aligned using computer programs.
- These sequences were subsequently annotated and were assigned to each chromosome.
Applications of HGP
- Biological systems can be well studied by the knowledge of DNA sequences.
- The Human Genome sequence was used to develop a new approach to biological research.
Also Read: Human Genome Project
DNA Fingerprinting
DNA fingerprinting involves identifying differences in some specific regions in DNA called repetitive DNA because, in these sequences, a small stretch of DNA is repeated many times. Alec Jeffrey who initially developed this technique used satellite DNA as the basis of DNA fingerprinting that shows a very high degree of polymorphism. It was called Variable Number Tandem Repeats. (VNTR). Its steps are;
- Isolation of DNA.
- Digestion of DNA by restriction endonucleases.
- Separation of DNA fragments by gel electrophoresis.
- Transferring (blotting) separated DNA fragments to synthetic membranes, such as nitrocellulose or nylon.
- Double-stranded DNA is made single-stranded by opening the bonds.
- Hybridization is done using a labeled VNTR probe.
- Detection of hybridized DNA fragments After hybridization with the VNTR probe the autoradiogram gives many bands of different sizes
- These bands give a characteristic pattern for an individual DNA. It differs from individual to individual.
The basis of DNA fingerprinting is;
- Repetitive DNA: DNA carries some non-coding repeated sequences.
- Satellite DNA: These are highly-repeated short sequences in the repetitive DNA.
Applications of DNA Fingerprinting
- Test of paternity.
- Identification of the criminals in unknown cases based on tissue samples e.g. rape, murder, etc.
- Population diversity determination or phylogenic status of animals
- Determination of genetic diseases.
FAQs on the Molecular Basis of Inheritance
Q1: Why DNA replication is semi-conservative?
Answer:
Because after the process of replication, the newly formed DNA will have one parental strand and one newly synthesized strand. As one parental strand is conserved, it is called semi-conservative replication.
Q2: In the process of transcription why both the strands are not copied into mRNA?
Answer:
Because if both strand act as a template then there is a possibility that two RNA molecules will be produced with different sequences. It results in the formation of 2 different proteins.
Q3: Why capping and tailing is required during post-transcriptional modification in eukaryotes?
Answer:
Capping and tailing are required because it checks the degradation of the mRNA by hydrolytic enzymes in the cytoplasm.
Like Article
Suggest improvement
Share your thoughts in the comments
Please Login to comment...