ML | Kaggle Breast Cancer Wisconsin Diagnosis using Logistic Regression
Last Updated :
01 Feb, 2022
Dataset :
It is given by Kaggle from UCI Machine Learning Repository, in one of its challenge
https://www.kaggle.com/uciml/breast-cancer-wisconsin-data. It is a dataset of Breast Cancer patients with Malignant and Benign tumor.
Logistic Regression is used to predict whether the given patient is having Malignant or Benign tumor based on the attributes in the given dataset.
Code : Loading Libraries
Python3
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
|
Code : Loading dataset
Python3
data = pd.read_csv("..\\breast-cancer-wisconsin-data\\data.csv")
print (data.head)
|
Output :
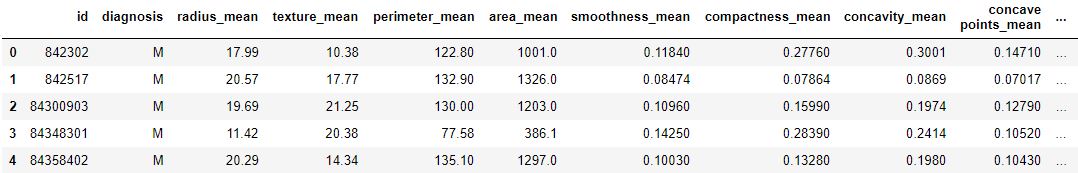
Code : Loading dataset
Output :
RangeIndex: 569 entries, 0 to 568
Data columns (total 33 columns):
id 569 non-null int64
diagnosis 569 non-null object
radius_mean 569 non-null float64
texture_mean 569 non-null float64
perimeter_mean 569 non-null float64
area_mean 569 non-null float64
smoothness_mean 569 non-null float64
compactness_mean 569 non-null float64
concavity_mean 569 non-null float64
concave points_mean 569 non-null float64
symmetry_mean 569 non-null float64
fractal_dimension_mean 569 non-null float64
radius_se 569 non-null float64
texture_se 569 non-null float64
perimeter_se 569 non-null float64
area_se 569 non-null float64
smoothness_se 569 non-null float64
compactness_se 569 non-null float64
concavity_se 569 non-null float64
concave points_se 569 non-null float64
symmetry_se 569 non-null float64
fractal_dimension_se 569 non-null float64
radius_worst 569 non-null float64
texture_worst 569 non-null float64
perimeter_worst 569 non-null float64
area_worst 569 non-null float64
smoothness_worst 569 non-null float64
compactness_worst 569 non-null float64
concavity_worst 569 non-null float64
concave points_worst 569 non-null float64
symmetry_worst 569 non-null float64
fractal_dimension_worst 569 non-null float64
Unnamed: 32 0 non-null float64
dtypes: float64(31), int64(1), object(1)
memory usage: 146.8+ KB
Code: We are dropping columns – ‘id’ and ‘Unnamed: 32’ as they have no role in prediction
Python3
data.drop(['Unnamed: 32', 'id'], axis = 1)
data.diagnosis = [1 if each == "M" else 0 for each in data.diagnosis]
|
Code : Input and Output data
Python3
y = data.diagnosis.values
x_data = data.drop(['diagnosis'], axis = 1)
|
Code : Normalisation
Python3
x = (x_data - np.min(x_data))/(np.max(x_data) - np.min(x_data)).values
|
Code : Splitting data for training and testing.
Python3
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size = 0.15, random_state = 42)
x_train = x_train.T
x_test = x_test.T
y_train = y_train.T
y_test = y_test.T
print("x train: ", x_train.shape)
print("x test: ", x_test.shape)
print("y train: ", y_train.shape)
print("y test: ", y_test.shape)
|
Code : Weight and bias
Python3
def initialize_weights_and_bias(dimension):
w = np.full((dimension, 1), 0.01)
b = 0.0
return w, b
|
Code : Sigmoid Function – calculating z value.
Python3
def sigmoid(z):
y_head = 1/(1 + np.exp(-z))
return y_head
|
Code : Forward-Backward Propagation
Python3
def forward_backward_propagation(w, b, x_train, y_train):
z = np.dot(w.T, x_train) + b
y_head = sigmoid(z)
loss = - y_train * np.log(y_head) - (1 - y_train) * np.log(1 - y_head)
cost = (np.sum(loss)) / x_train.shape[1]
derivative_weight = (np.dot(x_train, (
(y_head - y_train).T))) / x_train.shape[1]
derivative_bias = np.sum(
y_head-y_train) / x_train.shape[1]
gradients = {"derivative_weight": derivative_weight,
"derivative_bias": derivative_bias}
return cost, gradients
|
Code : Updating Parameters
Python3
def update(w, b, x_train, y_train, learning_rate, number_of_iterarion):
cost_list = []
cost_list2 = []
index = []
for i in range(number_of_iterarion):
cost, gradients = forward_backward_propagation(w, b, x_train, y_train)
cost_list.append(cost)
w = w - learning_rate * gradients["derivative_weight"]
b = b - learning_rate * gradients["derivative_bias"]
if i % 10 == 0:
cost_list2.append(cost)
index.append(i)
print ("Cost after iteration % i: % f" %(i, cost))
parameters = {"weight": w, "bias": b}
plt.plot(index, cost_list2)
plt.xticks(index, rotation ='vertical')
plt.xlabel("Number of Iterarion")
plt.ylabel("Cost")
plt.show()
return parameters, gradients, cost_list
|
Code : Predictions
Python3
def predict(w, b, x_test):
z = sigmoid(np.dot(w.T, x_test)+b)
Y_prediction = np.zeros((1, x_test.shape[1]))
for i in range(z.shape[1]):
if z[0, i]<= 0.5:
Y_prediction[0, i] = 0
else:
Y_prediction[0, i] = 1
return Y_prediction
|
Code : Logistic Regression
Python3
def logistic_regression(x_train, y_train, x_test, y_test,
learning_rate, num_iterations):
dimension = x_train.shape[0]
w, b = initialize_weights_and_bias(dimension)
parameters, gradients, cost_list = update(
w, b, x_train, y_train, learning_rate, num_iterations)
y_prediction_test = predict(
parameters["weight"], parameters["bias"], x_test)
y_prediction_train = predict(
parameters["weight"], parameters["bias"], x_train)
print("train accuracy: {} %".format(
100 - np.mean(np.abs(y_prediction_train - y_train)) * 100))
print("test accuracy: {} %".format(
100 - np.mean(np.abs(y_prediction_test - y_test)) * 100))
logistic_regression(x_train, y_train, x_test,
y_test, learning_rate = 1, num_iterations = 100)
|
Output :
Cost after iteration 0: 0.692836
Cost after iteration 10: 0.498576
Cost after iteration 20: 0.404996
Cost after iteration 30: 0.350059
Cost after iteration 40: 0.313747
Cost after iteration 50: 0.287767
Cost after iteration 60: 0.268114
Cost after iteration 70: 0.252627
Cost after iteration 80: 0.240036
Cost after iteration 90: 0.229543
Cost after iteration 100: 0.220624
Cost after iteration 110: 0.212920
Cost after iteration 120: 0.206175
Cost after iteration 130: 0.200201
Cost after iteration 140: 0.194860

Output :
train accuracy: 95.23809523809524 %
test accuracy: 94.18604651162791 %
Code : Checking results with linear_model.LogisticRegression
Python3
from sklearn import linear_model
logreg = linear_model.LogisticRegression(random_state = 42, max_iter = 150)
print("test accuracy: {} ".format(
logreg.fit(x_train.T, y_train.T).score(x_test.T, y_test.T)))
print("train accuracy: {} ".format(
logreg.fit(x_train.T, y_train.T).score(x_train.T, y_train.T)))
|
Output :
test accuracy: 0.9651162790697675
train accuracy: 0.9668737060041408
Like Article
Suggest improvement
Share your thoughts in the comments
Please Login to comment...