Mahotas – Highlighting Image Maxima
Last Updated :
17 May, 2022
In this article we will see how we can highlight the maxima of image in mahotas. Maxima can be best found in the distance map image because in labeled image each label is maxima but in distance map maxima can be identified easily. For this we are going to use the fluorescent microscopy image from a nuclear segmentation benchmark. We can get the image with the help of command given below
mahotas.demos.nuclear_image()
Below is the nuclear_image
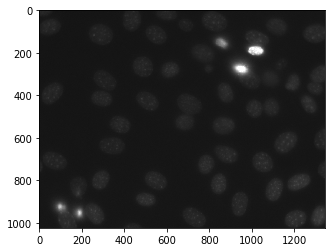
In order to do this we will use mahotas.morph.regmax method
Syntax : mahotas.morph.regmax(img, Bc)
Argument : It takes image object and numpy ones array as argument
Return : It returns image object
Note : The input of this should be the filtered image or loaded as grey
In order to filter the image we will take the image object which is numpy.ndarray and filter it with the help of indexing, below is the command to do this
image = image[:, :, 0]
Example 1 :
Python3
import mahotas
import mahotas.demos
import mahotas as mh
import numpy as np
from pylab import imshow, show
nuclear = mahotas.demos.nuclear_image()
nuclear = nuclear[:, :, 0]
nuclear = mahotas.gaussian_filter(nuclear, 4)
threshed = (nuclear > nuclear.mean())
dmap = mahotas.distance(threshed)
print("Distance Map")
imshow(dmap)
show()
Bc = np.ones((3, 2))
maxima = mahotas.morph.regmax(dmap, Bc = Bc)
print("Maxima")
imshow(maxima)
show()
|
Output :

Example 2 :
Python3
import numpy as np
import mahotas
from pylab import imshow, show
img = mahotas.imread('dog_image.png')
img = img[:, :, 0]
gaussian = mahotas.gaussian_filter(img, 15)
gaussian = (gaussian > gaussian.mean())
labelled, n_nucleus = mahotas.label(gaussian)
dmap = mahotas.distance(labelled)
print("Distance Map")
imshow(dmap)
show()
Bc = np.ones((4, 1))
maxima = mahotas.morph.regmax(dmap, Bc = Bc)
print("Maxima")
imshow(maxima)
show()
|
Output :

Like Article
Suggest improvement
Share your thoughts in the comments
Please Login to comment...