Ledoit-Wolf vs OAS Estimation in Scikit Learn
Last Updated :
25 Jan, 2023
Generally, Shrinkage is used to regularize the usual covariance maximum likelihood estimation. Ledoit and Wolf proposed a formula which is known as the Ledoit-Wolf covariance estimation formula; This close formula can compute the asymptotically optimal shrinkage parameter with minimizing a Mean Square Error(MSE) criterion feature. After that, one researcher Chen et al. made improvements in the Ledoit-Wolf Shrinkage parameter. He proposed Oracle Approximating Shrinkage (OAS) coefficient whose convergence is significantly better under the assumption that the data are Gaussian.
There are some basic concepts are needed to develop the Ledoit-Wolf estimators given below step by step:
Importing Libraries and Setting up the Variables
Python libraries make it very easy for us to handle the data and perform typical and complex tasks with a single line of code.
- Numpy – Numpy arrays are very fast and can perform large computations in a very short time.
- Matplotlib/Seaborn – This library is used to draw visualizations.
- Sklearn – This module contains multiple libraries having pre-implemented functions to perform tasks from data preprocessing to model development and evaluation.
- Scipy – SciPy is a python library that is useful in solving many mathematical equations and algorithms at its core it uses NumPy to handle the numbers.
Python3
import numpy as np
from sklearn.covariance import LedoitWolf, OAS
from scipy.linalg import toeplitz, cholesky
import matplotlib.pyplot as plt
np.random.seed(0)
numOffeatures = 250
r = 0.25
realTimeCovariance = toeplitz(r ** np.arange(numOffeatures))
colorMatrix = cholesky(realTimeCovariance)
|
Creating Variables to Store Results
Now we will initialize some NumPy matrices so, that we can store the results from the estimation to them. Here, the mean squared error calculation logic and shrinkage estimation logic of both Ledoit-Wolf and OAS is being specified.
Python3
numOfSamplesRange = np.arange(8, 40, 1)
repeat = 150
ledoitWolf_mse = np.zeros((numOfSamplesRange.size, repeat))
oas_mse = np.zeros((numOfSamplesRange.size, repeat))
ledoitWolf_shrinkage = np.zeros((numOfSamplesRange.size, repeat))
oas_shrinkage = np.zeros((numOfSamplesRange.size, repeat))
|
Parameters
- Store_precision: bool, default=True
- Assume_centered: bool, default=False
- Block_size: int, default=1000
Attributes
- Covariance_: array of shape (n_features, n_features)
- Precision_: array of shape (n_features, n_features)
- shrinkage_: float
- n_features_in_: int
Now, we will calculate the MSE and shrinkage by using nested for loop to fit the values for both models step by step:
Python3
for i, numOfSamples in enumerate(numOfSamplesRange):
for j in range(repeat):
X = np.dot(np.random.normal(size=(numOfSamples, numOffeatures)),
colorMatrix.T)
ledoitWolf = LedoitWolf(store_precision=False,
assume_centered=True)
ledoitWolf.fit(X)
ledoitWolf_mse[i, j] = ledoitWolf.error_norm(realTimeCovariance,
scaling=False)
ledoitWolf_shrinkage[i, j] = ledoitWolf.shrinkage_
oas = OAS(store_precision=False, assume_centered=True)
oas.fit(X)
oas_mse[i, j] = oas.error_norm(realTimeCovariance,
scaling=False)
oas_shrinkage[i, j] = oas.shrinkage_
|
Visualizing MSE and Shrinkage
Now let’s create a graph for both of the estimations first for error and then for shrinkage. Here, graphs for the comparison between Ledoit-Wolf and OAS have been plotted in the factor of MSE and shrinkage accordingly.
Python3
plt.subplot(2, 1, 1)
plt.errorbar(
numOfSamplesRange,
ledoitWolf_mse.mean(1), yerr=ledoitWolf_mse.std(1),
label="Ledoit-Wolf", color="yellow", lw=2,
)
plt.errorbar(
numOfSamplesRange,
oas_mse.mean(1), yerr=oas_mse.std(1),
label="OAS", color="green", lw=2,
)
plt.ylabel("Squared error")
plt.legend(loc="upper right")
plt.title("Comparison of covariance estimators(Ledoit-Wolf vs OAS)")
plt.xlim(2, 32)
plt.show()
|
Output:
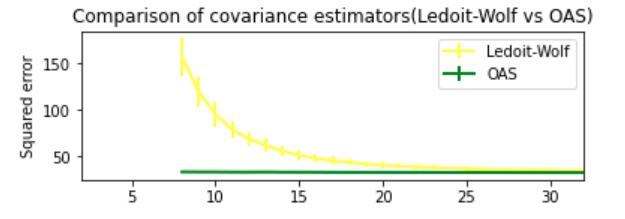
Ledoit-Wolf vs OAS (comparison factor is MSE)
Python3
plt.subplot(2, 1, 2)
plt.errorbar(
numOfSamplesRange,
ledoitWolf_shrinkage.mean(1), yerr=ledoitWolf_shrinkage.std(1),
label="Ledoit-Wolf", color="yellow", lw=2,
)
plt.errorbar(
numOfSamplesRange,
oas_shrinkage.mean(1), yerr=oas_shrinkage.std(1),
label="OAS", color="green", lw=2,
)
plt.xlabel("number of samples")
plt.ylabel("Shrinkage counting")
plt.legend(loc="lower right")
plt.ylim(plt.ylim()[0], 1.0 + (plt.ylim()[1] - plt.ylim()[0]) / 10.0)
plt.xlim(2, 32)
plt.show()
|
Output:
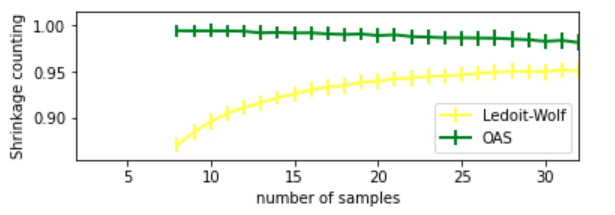
Ledoit-Wolf vs OAS (comparison factor is shrinkage)
Like Article
Suggest improvement
Share your thoughts in the comments
Please Login to comment...