Gradient Descent With RMSProp from Scratch
Last Updated :
08 Jun, 2023
Gradient descent is an optimization algorithm used to find the set of parameters (coefficients) of a function that minimizes a cost function. This method iteratively adjusts the coefficients of the function until the cost reaches the local, or global, minimum. Gradient descent works by calculating the partial derivatives of the cost function with respect to each of the coefficients. The algorithm then makes small adjustments to the coefficients in order to reduce the cost until it reaches a minimum. The direction of the adjustments is determined by the negative of the gradient of the cost function. Optimizers are methods or algorithms that reduce a loss (an error) by adjusting various parameters and weights, minimizing the loss function, and thereby improving model accuracy and speed. One such optimization technique is RMSprop.
RMSProp (Root Mean Squared Propagation) is an adaptive learning rate optimization algorithm. It is an extension of the popular Adaptive Gradient Algorithm and is designed to dramatically reduce the amount of computational effort used in training neural networks. This algorithm works by exponentially decaying the learning rate every time the squared gradient is less than a certain threshold. This helps reduce the learning rate more quickly when the gradients become small. In this way, RMSProp is able to smoothly adjust the learning rate for each of the parameters in the network, providing a better performance than regular Gradient Descent alone.
The RMSprop algorithm utilizes exponentially weighted moving averages of squared gradients to update the parameters. Here is the mathematical equation for RMSprop:
- Initialize parameters:
- Learning rate: α
- Exponential decay rate for averaging: γ
- Small constant for numerical stability: ε
- Initial parameter values: θ
- Initialize accumulated gradients (Exponentially weighted average):
- Accumulated squared gradient for each parameter: Et= 0
- Repeat until convergence or maximum iterations:
- Compute the gradient of the objective function with respect to the parameters:
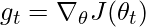
- Update the exponentially weighted average of the squared gradients:
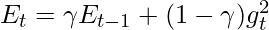
- Update the parameters:
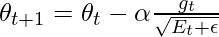
where,
- gt is the gradient of the loss function with respect to the parameters at time t
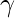 is a decay factor
is a decay factor- Et is the exponentially weighted average of the squared gradients
- α is the learning rate
- ϵ is a small constant to prevent division by zero
This process is repeated for each parameter in the optimization problem, and it helps adjust the learning rate for each parameter based on the historical gradients. The exponential moving average allows the algorithm to give more importance to recent gradients and dampen the effect of older gradients, providing stability during optimization.
Implementation
Now, we will look into the implementation of the RMSprop. We will first import all the necessary libraries as follows.
Python3
import numpy as np
import matplotlib.pyplot as plt
from numpy import arange, meshgrid
|
Now, we will define our objective function and its derivatives. For this article we are considering the objective function to be 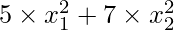 where x1 and x2 are variables.
where x1 and x2 are variables.
Python3
def objective(x1, x2):
return 5 * x1**2.0 + 7 * x2**2.0
def derivative_x1(x1, x2):
return 10.0 * x1
def derivative_x2(x1, x2):
return 14.0 * x2
|
Now, let us visualize this equation. We will look into its 3D graph between x1, x2, and y and we will also look into its 2D representation (contour plot).
Python3
x1 = arange(-5.0, 5.0, 0.1)
x2 = arange(-5.0, 5.0, 0.1)
x1, x2 = meshgrid(x1, x2)
y = objective(x1, x2)
fig = plt.figure(figsize=(12, 4))
ax = fig.add_subplot(1, 2, 1, projection='3d')
ax.plot_surface(x1, x2, y, cmap='viridis')
ax.set_xlabel('x1')
ax.set_ylabel('x2')
ax.set_zlabel('y')
ax.set_title('3D plot of the objective function')
ax = fig.add_subplot(1, 2, 2)
ax.contour(x1, x2, y, cmap='viridis', levels=20)
ax.set_xlabel('x1')
ax.set_ylabel('x2')
ax.set_title('Contour plot of the objective function')
plt.show()
|
Output:
-(2).png)
Now, let us define our RMSprop optimizer.
Python3
def rmsprop(x1, x2, derivative_x1, derivative_x2, learning_rate, gamma, epsilon, max_epochs):
x1_trajectory = []
x2_trajectory = []
y_trajectory = []
x1_trajectory.append(x1)
x2_trajectory.append(x2)
y_trajectory.append(objective(x1, x2))
e1 = 0
e2 = 0
for _ in range(max_epochs):
gt_x1 = derivative_x1(x1, x2)
gt_x2 = derivative_x2(x1, x2)
e1 = gamma * e1 + (1 - gamma) * gt_x1**2.0
e2 = gamma * e2 + (1 - gamma) * gt_x2**2.0
x1 = x1 - learning_rate * gt_x1 / (np.sqrt(e1 + epsilon))
x2 = x2 - learning_rate * gt_x2 / (np.sqrt(e2 + epsilon))
x1_trajectory.append(x1)
x2_trajectory.append(x2)
y_trajectory.append(objective(x1, x2))
return x1_trajectory, x2_trajectory, y_trajectory
|
Now, let us optimize our objective function using the RMSprop function.
Python3
x1_initial = -4.0
x2_initial = 3.0
learning_rate = 0.1
gamma = 0.9
epsilon = 1e-8
max_epochs = 50
x1_trajectory, x2_trajectory, y_trajectory = rmsprop(
x1_initial,
x2_initial,
derivative_x1,
derivative_x2,
learning_rate,
gamma,
epsilon,
max_epochs
)
print('The optimal value of x1 is:', x1_trajectory[-1])
print('The optimal value of x2 is:', x2_trajectory[-1])
print('The optimal value of y is:', y_trajectory[-1])
|
Output:
The optimal value of x1 is: -0.10352260359924752
The optimal value of x2 is: 0.0025296212056016548
The optimal value of y is: 0.05362944016394148
Now, let us visualize the path or trajectory of the objective function.
Python3
fig = plt.figure(figsize=(6, 6))
ax = fig.add_subplot(1, 1, 1)
ax.contour(x1, x2, y, cmap='viridis', levels=20)
ax.plot(x1_trajectory, x2_trajectory, '*',
markersize=7, color='dodgerblue')
ax.set_xlabel('x1')
ax.set_ylabel('x2')
ax.set_title('RMSprop Optimization path for ' + str(max_epochs) + ' iterations')
plt.show()
|
Output:
.png)
Like Article
Suggest improvement
Share your thoughts in the comments
Please Login to comment...