Exploratory Data Analysis or EDA is a statistical approach or technique for analyzing data sets to summarize their important and main characteristics generally by using some visual aids. The EDA approach can be used to gather knowledge about the following aspects of data.
- Main characteristics or features of the data.
- The variables and their relationships.
- Finding out the important variables that can be used in our problem.
EDA is an iterative approach that includes:
- Generating questions about our data
- Searching for the answers by using visualization, transformation, and modeling of our data.
- Using the lessons that we learn to refine our set of questions or to generate a new set of questions.
Exploratory Data Analysis in R
In R Programming Language, we are going to perform EDA under two broad classifications:
- Descriptive Statistics, which includes mean, median, mode, inter-quartile range, and so on.
- Graphical Methods, which includes histogram, density estimation, box plots, and so on.
Before we start working with EDA, we must perform the data inspection properly. Here in our analysis, we will be using the loafercreek from the soilDB package in R. We are going to inspect our data in order to find all the typos and blatant errors. Further EDA can be used to determine and identify the outliers and perform the required statistical analysis. For performing the EDA, we will have to install and load the following packages:
- “aqp” package
- “ggplot2” package
- “soilDB” package
We can install these packages from the R console using the install.packages() command and load them into our R Script by using the library() command. We will now see how to inspect our data and remove the typos and blatant errors.
Data Inspection for Exploratory Analysis in R
To ensure that we are dealing with the right information we need a clear view of your data at every stage of the transformation process. Data Inspection is the act of viewing data for verification and debugging purposes, before, during, or after a translation. Now let’s see how to inspect and remove the errors and typos from the data.
R
# Data Inspection in EDA
# loading the required packages
library(aqp)
library(soilDB)
# Load from the loafercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and
# add to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname,
n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing of
# the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
Output:
2BC 2BCt 2Bt1 2Bt2 2Bt3 2Bt4 2Bt5 2CB 2CBt 2Cr 2Crt 2R A A1 A2 AB
A 0 0 0 0 0 0 0 0 0 0 0 0 97 7 7 0
BAt 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1
Bt1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Bt2 1 1 3 8 8 6 1 1 1 0 0 0 0 0 0 0
Cr 0 0 0 0 0 0 0 0 0 4 2 0 0 0 0 0
R 0 0 0 0 0 0 0 0 0 0 0 6 0 0 0 0
not-used 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
ABt Ad Ap B BA BAt BC BCt Bt Bt1 Bt2 Bt3 Bt4 Bw Bw1 Bw2 Bw3 C
A 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
BAt 0 0 0 0 31 8 0 0 0 0 0 0 0 0 0 0 0 0
Bt1 2 0 0 0 0 0 0 0 8 93 88 0 0 10 2 2 1 0
Bt2 0 0 0 0 0 0 4 16 0 0 0 47 8 0 0 0 0 6
Cr 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
R 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
not-used 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0
CBt Cd Cr Cr/R Crt H1 Oi R Rt
A 0 0 0 0 0 0 0 0 0
BAt 0 0 0 0 0 0 0 0 0
Bt1 0 0 0 0 0 0 0 0 0
Bt2 6 1 0 0 0 0 0 0 0
Cr 0 0 49 0 20 0 0 0 0
R 0 0 0 1 0 0 0 40 1
not-used 0 0 0 0 0 1 24 0 0
genhz clay total_frags_pct phfield
A :113 Min. :10.00 Min. : 0.00 Min. :4.90
BAt : 40 1st Qu.:18.00 1st Qu.: 0.00 1st Qu.:6.00
Bt1 :206 Median :22.00 Median : 5.00 Median :6.30
Bt2 :118 Mean :23.63 Mean :13.88 Mean :6.18
Cr : 75 3rd Qu.:28.00 3rd Qu.:20.00 3rd Qu.:6.50
R : 48 Max. :60.00 Max. :95.00 Max. :7.00
not-used: 26 NA's :167 NA's :381
effclass
Length:626
Class :character
Mode :character
[1] "2BC" "2BCt" "2Bt1" "2Bt2" "2Bt3" "2Bt4" "2Bt5" "2CB" "2CBt" "2Cr"
[11] "2Crt" "2R" "A" "A1" "A2" "AB" "ABt" "Ad" "Ap" "B"
[21] "BA" "BAt" "BC" "BCt" "Bt" "Bt1" "Bt2" "Bt3" "Bt4" "Bw"
[31] "Bw1" "Bw2" "Bw3" "C" "CBt" "Cd" "Cr" "Cr/R" "Crt" "H1"
[41] "Oi" "R" "Rt"
Now proceed with the EDA.
Descriptive Statistics Exploratory Data Analysis in R
For Descriptive Statistics in order to perform EDA in R, we will divide all the functions into the following categories:
- Measures of central tendency
- Measures of dispersion
- Correlation
We will try to determine the mid-point values using the functions under the Measures of Central tendency. Under this section, we will be calculating the mean, median, mode, and frequencies.
The measures of central tendency
R
# first remove missing values
# and create a new vector
clay <- na.exclude(h$clay)
mean(clay)
median(clay)
sort(table(round(h$clay)),
decreasing = TRUE)[1]
table(h$genhz)
# append the table with
# row and column sums
addmargins(table(h$genhz,
h$texcl))
Output:
mean(clay)
[1] 23.62767
median(clay)
[1] 22
25
42
A BAt Bt1 Bt2 Cr R not-used
113 40 206 118 75 48 26
c cl l scl sic sicl sil sl Sum
A 0 0 78 0 0 0 27 6 111
BAt 0 2 31 0 0 1 4 1 39
Bt1 2 44 127 4 1 5 20 1 204
Bt2 16 54 29 5 1 3 5 0 113
Cr 1 0 0 0 0 0 0 0 1
R 0 0 0 0 0 0 0 0 0
not-used 0 1 0 0 0 0 0 0 1
Sum 19 101 265 9 2 9 56 8 469
Calculate the proportions
R
# relative to the rows, margin = 1
# calculates for rows, margin = 2 calculates
# for columns, margin = NULL calculates
# for total observations
round(prop.table(table(h$genhz, h$texture_class),
margin = 1) * 100)
knitr::kable(addmargins(table(h$genhz, h$texcl)))
aggregate(clay ~ genhz, data = h, mean)
aggregate(clay ~ genhz, data = h, median)
aggregate(clay ~ genhz, data = h, summary)
Output:
| | c| cl| l| scl| sic| sicl| sil| sl| Sum|
|:--------|--:|---:|---:|---:|---:|----:|---:|--:|---:|
|A | 0| 0| 78| 0| 0| 0| 27| 6| 111|
|BAt | 0| 2| 31| 0| 0| 1| 4| 1| 39|
|Bt1 | 2| 44| 127| 4| 1| 5| 20| 1| 204|
|Bt2 | 16| 54| 29| 5| 1| 3| 5| 0| 113|
|Cr | 1| 0| 0| 0| 0| 0| 0| 0| 1|
|R | 0| 0| 0| 0| 0| 0| 0| 0| 0|
|not-used | 0| 1| 0| 0| 0| 0| 0| 0| 1|
|Sum | 19| 101| 265| 9| 2| 9| 56| 8| 469|
genhz clay
1 A 16.23832
2 BAt 19.49730
3 Bt1 24.03166
4 Bt2 31.20783
5 Cr 15.00000
genhz clay
1 A 16
2 BAt 19
3 Bt1 24
4 Bt2 30
5 Cr 15
genhz clay.Min. clay.1st Qu. clay.Median clay.Mean clay.3rd Qu. clay.Max.
1 A 10.00000 14.00000 16.00000 16.23832 18.00000 25.00000
2 BAt 14.00000 17.00000 19.00000 19.49730 20.00000 28.00000
3 Bt1 12.00000 20.00000 24.00000 24.03166 27.00000 51.40000
4 Bt2 10.00000 26.00000 30.00000 31.20783 35.00000 60.00000
5 Cr 15.00000 15.00000 15.00000 15.00000 15.00000 15.00000
Now we will see the functions under Measures of Dispersion. In this category, we are going to determine the spread values around the mid-point. Here we are going to calculate the variance, standard deviation, range, inter-quartile range, coefficient of variance, and quartiles.
The measures of Dispersion
R
# first remove missing values
# and create a new vector
clay <- na.exclude(h$clay)
var(h$clay, na.rm=TRUE)
sd(h$clay, na.rm = TRUE)
cv <- sd(clay) / mean(clay) * 100
cv
quantile(clay)
range(clay)
IQR(clay)
Output:
var(h$clay, na.rm=TRUE)
[1] 64.89187
sd(h$clay, na.rm = TRUE)
[1] 8.055549
cv
[1] 34.03087
quantile(clay)
0% 25% 50% 75% 100%
10 18 22 28 60
range(clay)
[1] 10 60
IQR(clay)
[1] 10
Now we will work on Correlation. In this part, all the calculated correlation coefficient values of all variables in tabulated as the Correlation Matrix. This gives us a quantitative measure in order to guide our decision-making process.
We shall now see the correlation in this example.
R
# Compute the middle horizon depth
h$hzdepm <- (h$hzdepb + h$hzdept) / 2
vars <- c("hzdepm", "clay", "sand",
"total_frags_pct", "phfield")
round(cor(h[, vars], use = "complete.obs"), 2)
Output:
hzdepm clay sand total_frags_pct phfield
hzdepm 1.00 0.59 -0.08 0.50 -0.03
clay 0.59 1.00 -0.17 0.28 0.13
sand -0.08 -0.17 1.00 -0.05 0.12
total_frags_pct 0.50 0.28 -0.05 1.00 -0.16
phfield -0.03 0.13 0.12 -0.16 1.00
Hence, the above three classifications deal with the Descriptive statistics part of EDA. Now we shall move on to the Graphical Method of representing EDA.
Graphical Method in Exploratory Data Analysis in R
Since we have already checked our data for missing values, blatant errors, and typos, we can now examine our data graphically in order to perform EDA. We will see the graphical representation under the following categories:
- Distributions
- Scatter and Line plot
Under the Distribution, we shall examine our data using the bar plot, Histogram, Density curve, box plots, and QQplot.
We shall see how distribution graphs can be used to examine data in EDA in this example.
R
# Load necessary libraries
library(ggplot2)
library(aqp)
library(soilDB)
# Load from the loafercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and add
# to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname, n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing
# of the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
# Clean up hzname for better plotting
h$hzname <- ifelse(h$hzname == "BT", "Bt", h$hzname)
# Bar plot with color
ggplot(h, aes(x = reorder(texcl, clay), fill = texcl)) +
geom_bar() +
labs(title = "Bar Plot of Texture Class",
x = "Texture Class",
y = "Count") +
theme_minimal() +
theme(legend.position = "none")
Output:
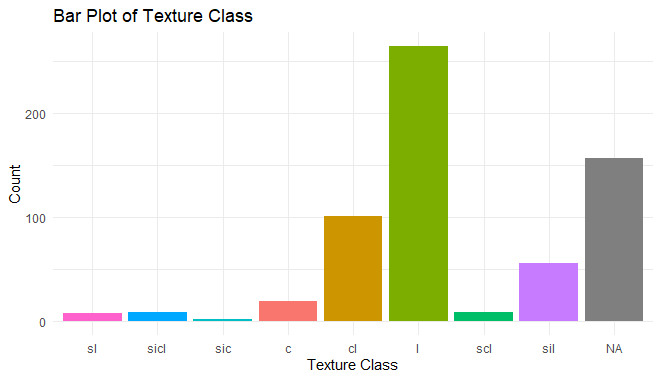
Exploratory Data Analysis in R Programming
Histogram with customized bins and color
R
# Histogram with customized bins and color
ggplot(h, aes(x = clay, fill = texcl)) +
geom_histogram(bins = 30, color = "white", alpha = 0.7) +
labs(title = "Histogram of Clay Content",
x = "Clay Content (%)",
y = "Frequency") +
theme_minimal()
Output:

Exploratory Data Analysis in R Programming
Density curve with fill color
R
# Density curve with fill color
ggplot(h, aes(x = clay, fill = texcl)) +
geom_density(alpha = 0.7) +
labs(title = "Density Curve of Clay Content",
x = "Clay Content (%)",
y = "Density") +
theme_minimal()
Output:

Exploratory Data Analysis in R Programming
Box plot with color and jitter
R
# Box plot with color and jitter
ggplot(h, aes(x = genhz, y = clay, fill = texcl)) +
geom_boxplot(show.legend = FALSE) +
geom_jitter(position = position_jitter(0.2), alpha = 0.7, color = "gray") +
labs(title = "Box Plot of Clay Content by Generalized Horizon",
x = "Generalized Horizon",
y = "Clay Content (%)") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
Output:
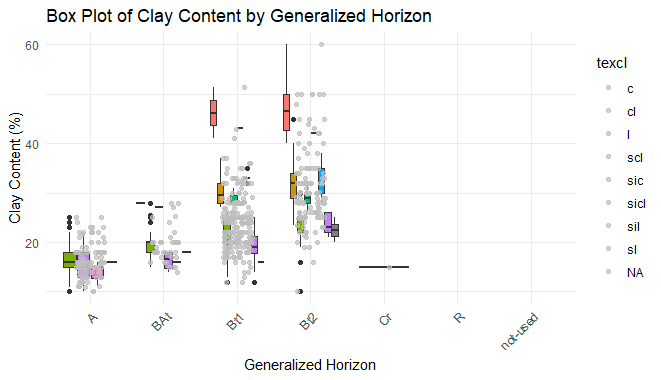
Exploratory Data Analysis in R Programming
QQ Plot for Clay
R
# QQ Plot for Clay
ggplot(h, aes(sample = clay)) +
geom_qq_line(color = "blue") +
geom_qq(color = "red", alpha = 0.7) +
labs(title = "QQ Plot for Clay Content",
x = "Theoretical Quantiles",
y = "Sample Quantiles") +
theme_minimal()
Output:
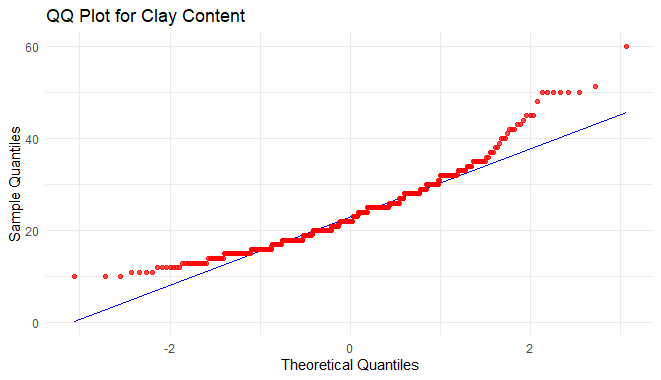
Exploratory Data Analysis in R Programming
Now we will move on to the Scatter and Line plot. In this category, we are going to see two types of plotting,- scatter plot and line plot. Plotting points of one interval or ratio variable against variable are known as a scatter plot.
We shall now see how to use scatter plot to examine our data.
R
# Extract the horizon table
h <- horizons(loafercreek)
# Scatter plot
ggplot(h, aes(x = total_frags_pct, y = clay, color = texcl)) +
geom_point(alpha = 0.7, size = 3) +
labs(title = "Scatter Plot of Total Fragments vs Clay Content",
x = "Total Fragments (%)",
y = "Clay Content (%)",
color = "Texture Class") +
theme_minimal()
Output:

Exploratory Data Analysis in R Programming
Conclusion
Exploratory Data Analysis (EDA) is a crucial phase in the data analysis process that empowers data scientists and analysts to gain insights into their datasets. In R programming, the combination of powerful libraries such as ggplot2, aqp, and others facilitates the creation of visually appealing and informative visualizations for EDA.
Like Article
Suggest improvement
Share your thoughts in the comments
Please Login to comment...