Biopython – Pairwise Alignment
Last Updated :
28 Jul, 2022
Pairwise Sequence Alignment is a process in which two sequences are compared at a time and the best possible sequence alignment is provided. Pairwise sequence alignment uses a dynamic programming algorithm. Biopython has a special module Bio.pairwise2 which identifies the alignment sequence using pairwise method. Biopython provides the best algorithm to find alignment sequence as compared to other software.
Let’s take two simple and hypothetical sequences as an example for using pairwise module.
Python3
from Bio import pairwise2
from Bio.Seq import Seq
seq1 = Seq("TGTGACTA")
seq2 = Seq("CATGGTCA")
alignments = pairwise2.align.globalxx(seq1, seq2)
for match in alignments:
print(match)
|
Output:
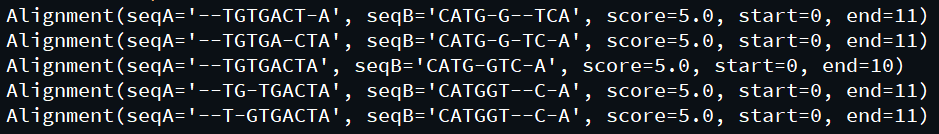
Here, the globalxx method does the main work, it follows the convention <alignment type>XX where XX is a code having two characters indicating the parameters it takes. First character indicates the matching and mismatching score while the second indicates the parameter for the gap penalty.
Match parameters :
| Code Character |
Description |
| x |
No parameters. Identical character has score of 1, else 0. |
| m |
match score of identical chars, else mismatch score. |
| d |
dictionary returning scores of any pair of characters. |
| c |
A callback function returns scores. |
gap penalty parameters :
| Code Character |
Description |
| x |
No gap penalties. |
| s |
both sequences having same open and extend gap penalty. |
| d |
sequences having different open and extend gap penalty. |
| c |
A callback function returns the gap penalties. |
For a nice printout Bio.pairwise2 provides format_alignment() method:
Python3
from Bio import pairwise2
from Bio.Seq import Seq
from Bio.pairwise2 import format_alignment
seq1 = Seq("TGTGACTA")
seq2 = Seq("CATGGTCA")
alignments = pairwise2.align.globalxx(seq1, seq2)
for alignment in alignments:
print(format_alignment(*alignment))
|
Output:
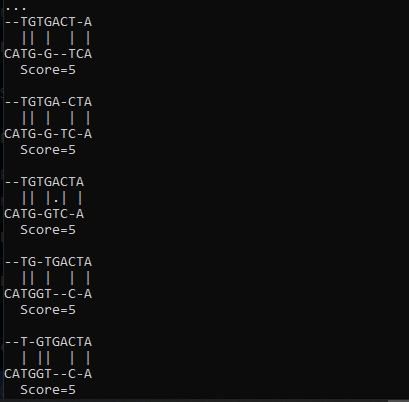
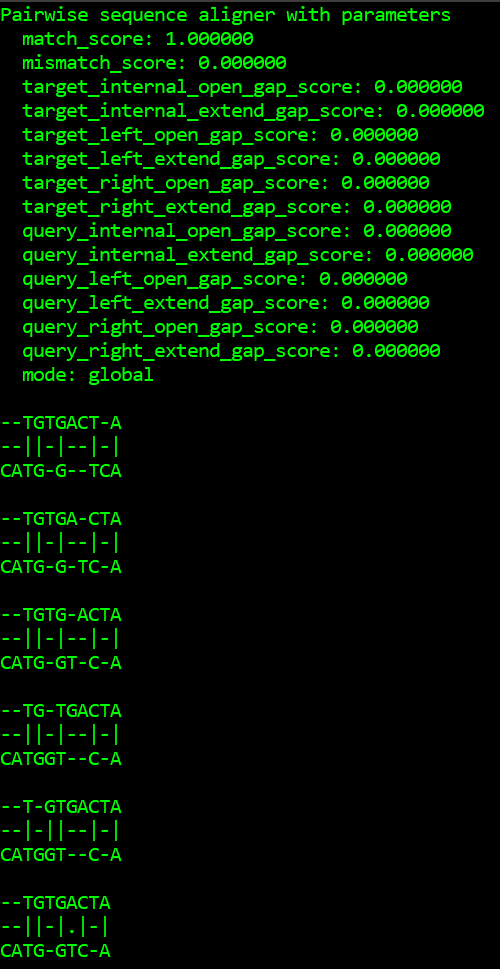
There is another module provided by Biopython to do the pairwise sequence alignment. Align module has a PairwiseAligner() for this purpose. It has various APIs to set the parameters like mode, match score, algorithm, gap penalty, etc. Below is a simple implementation of the method:
Python3
from Bio import Align
from Bio.Seq import Seq
seq1 = Seq("TGTGACTA")
seq2 = Seq("CATGGTCA")
aligner = Align.PairwiseAligner()
print(aligner)
alignments = aligner.align(seq1, seq2)
for alignment in alignments:
print(alignment)
|
Output:
Like Article
Suggest improvement
Share your thoughts in the comments
Please Login to comment...